JSBB: Volume 2, Issue 1, April 2023 - BIO-DATABASES, STRUCTURE & FUNCTION
Research article: Revisited research
The NALD database: A Comprehensive Resource for Understanding Molecular Interactions
RACHEDI Abdelkrim
Laboratory of Biotoxicology, Pharmacognosy and biological valorisation of plants, Faculty of Sciences, Department of Biology, University of Saida - Dr Moulay Tahar, 20100 Saida, Algeria.
📧 E. mail: abdelkrim.rachedi@univ-saida.dz
Published: 01 April 2023
Abstract
The Nucleic Acids and Ligands Database (NALD) is a comprehensive resource that provides
information on the interactions between nucleic acids and ligands. This paper describes
the development of NALD and its features, including the types of data it contains and
the user interface. NALD is a valuable resource for researchers in the fields of
molecular biology, biochemistry, and drug design.
The NALD database is concerned with the identification of ligands (drugs) that bind
nucleic acids (NA) and provide users with a verity of binding information existing between both molecules.
The database annotates nucleic acids in complexes with drugs in terms of detailed binding interactions,
binding motifs where binding occurs, binding properties, binding modes and classes and links to diseases.
These were calculated from entries of NA/Ligand complexes from the protein data bank (PDB) and also extracted
by both automatic and manual means from scientific literature sources such as the PubMed web site (PMID) and
publications. NALD provides online access to these types of information while it focuses on ligands that bind nucleic
acids with implications on diseases of high prevalence such as HIV/AIDS, cancer, hepatitis, malaria and tuberculosis.
This paper is a revisit of the research paper NALD: Nucleic Acids and Ligands Database (Rachedi & Madida, 2013).
The revisit highlights new improvements to the database including DNA/RNA-ligands binding motifs data growth, data-integration
and the implementation of a customized 3D-graphics tool using the PDBe-RCSB Mol* (Sehnal D. et. al., 2021).
Availability
The NALD database is available online from the University of Saida, Algeria, via the URL address:
https://bioinformatics.univ-saida.dz/bit2/?arg=SB1
Key words
Bioinformatics, Database, Data mining, Data integration, Nucleic
acids, Binding motifs, Ligands, Drugs, Diseases
Introduction
The interactions between nucleic acids and ligands play a crucial role in various biological processes,
such as gene expression and DNA replication. Understanding the nature of these interactions is essential
for the development of new drugs and therapies. To facilitate research in this area,
Nucleic Acids and Ligands Database (NALD) was developed as a comprehensive resource that provides information on the
molecular interactions between nucleic acids and ligands.
Nucleic acid molecules are biologically important found in all living organisms. They contain genetic material that must be
synthesized and reproduced with high fidelity to ensure its proper function. Failure to do so conditions of compromised health and
disease arise and for this reason nucleic acids are points of interest for therapeutic drug targets during specific binding events.
The protein data bank (PDB) (Berman et al., 2000) is an international repository of a large number of 3D structures for macromolecules
including protein, nucleic acids and their complexes.
Many of the compounds binding nucleic acids known in general as ligands are considered drugs either designed as therapeutics or
as additives for structural and functional investigation. This project deals with the development of an online database, Nucleic
Acids and Ligands Database (NALD), which annotates the nucleic acid (NA) in complexes with ligand (drugs) in terms of detailed
binding interactions, NA motifs where binding occurs, binding properties, binding modes and classes and links to diseases. Focus
will be centered on ligands that bind nucleic acids with implications on diseases of high prevalence region such as HIV/AIDS, cancer,
hepatitis, malaria and tuberculosis. The database provide data integration in links to the PDB, PDBeChem (Dimitropoulos et.al., 2006)
and other literature resources such as UniProt and PubMed databases.
Methods
NALD was developed by collecting data from various sources, including the Protein Data Bank (PDB),
the Cambridge Structural Database (CSD), and the literature. The data was then curated and organized
into a user-friendly database that provides detailed information on the interactions between nucleic
acids and ligands.
The database consists of two models of information storage; MySql Relational Module and FlaFiles
modules. The relational module,
created in MySQL database platfrom, is where tables, governed by a database schema, are used to store related information and the
linking of the table components is via unique identifier columns or primary keys. The second module, flatfile
structure, stores
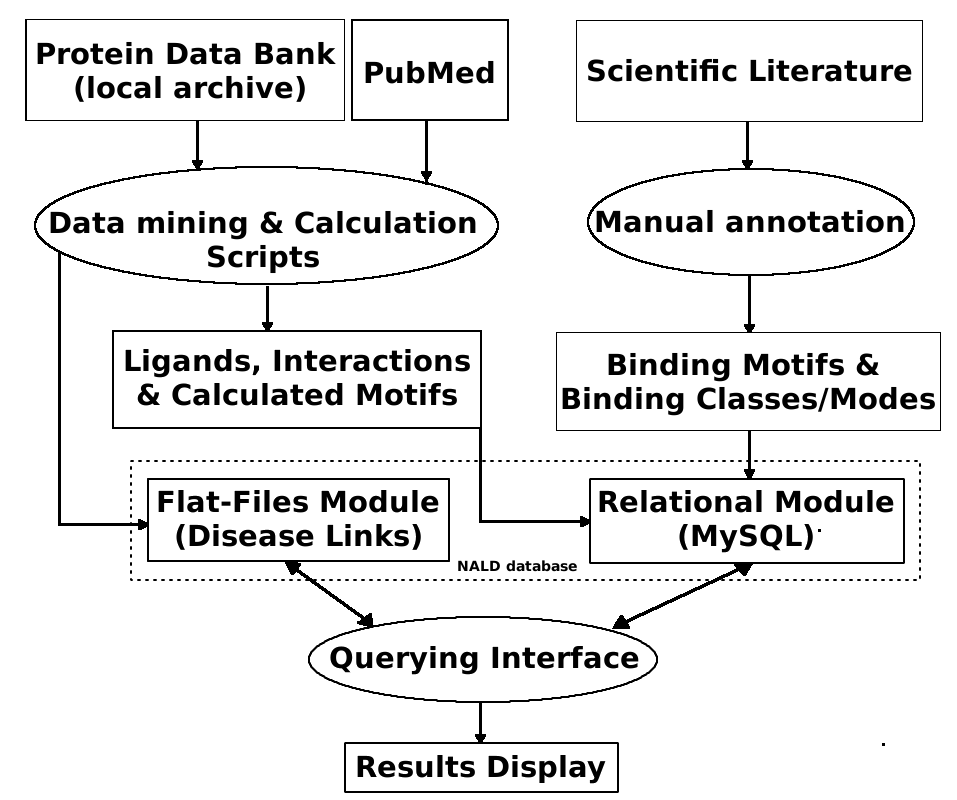
information in plain text files, Figure 1.
Figure 1. Diagram description of components that make the NALD database. Data is mined from the PDB and PubMed using PHP scripts and conduct
calculation of NA/Ligands interaction, calculated binding motifs. Scientific literature is also used for manual extraction of empirical information about
NA/Ligands binding motifs and binding binding and properties. The data are then stored into a MySQL based tables. Information about disease links to
ligands which is extracted automatically from PubMed is stored in flatfiles
format.
The tables present in MySQL were created using PHP scripts that acquire data and simultaneously create and loaded information
directly into the MySQL tables. The tables were populated by the data mined from the Ligands Sites Explorer (LSE) website which is a
locally developed online system that uses an archive version of the Protein Data Bank (PDB) data. The tables are created with a
hierarchical structural component from the PDBid table cascading down to the classes of the ligands. The information pertaining to
calculated binding motifs and ligand interactions with nucliec acids are based on exclusively complexes of nucleic acids (DNA, RNA
and hybrids) with ligands as found in the PDB. Data related to binding motifs, binding modes/classes and properties have been
manually mined from scientific literature and then loaded in MysSQL tables. Disease links data have been mined from the PubMed
database.
Results
The database is updated on regular basis and currently contain 2258 NA/Ligands complexes and 917 unique ligands, Figure 3C.
Annotated classes of NA–Ligands binding covers Intercalation, Modification, Addition and Cavity fitting in addition to binding
modes of ligands to the DNA's Minor Groove, Major Groove and both Minor/Major Grooves. Annotation included binding motifs, binding sites
and properties (Figure 3B). In this late version of the NALD database, the PDBe-RCSB Mol* (Sehnal D. et. al., 2021) customized 3D graphics is implemented and used to illustrate the
binding of ligands to their NA targets, Figure 4D.
Querying the NALD database:
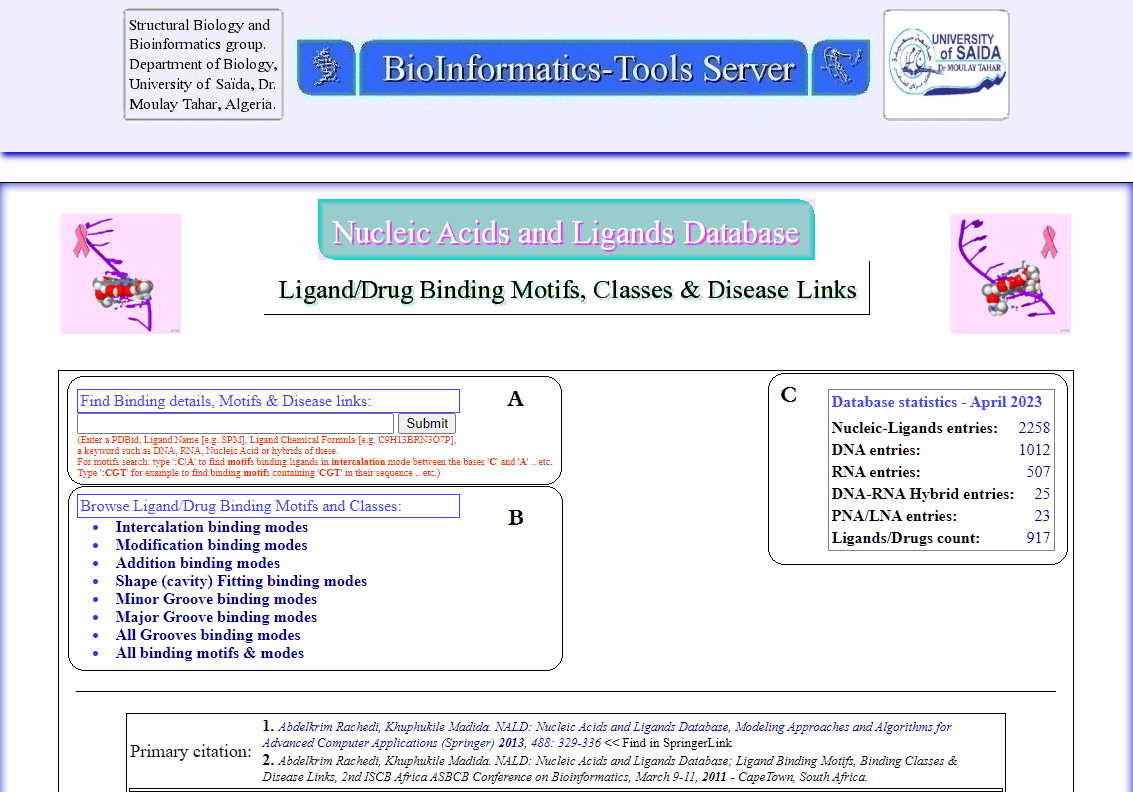
NALD database offers two ways for data retrieval; “Find Binding details & Disease links:” which is a search box, Figure 2A., and
“Browse Ligand/Drug Binding Motifs and Classes:”, which offers links for details of modes of binding, Figure 2B.
Figure 2. Screenshot from NALD main interface. A. search box for information retrieval; “Find Binding details & Disease links:” and B. Browse Ligand/Drug Binding Motifs and Classes:. C. Monthly updated statistics of the database content.
Find Binding details & Disease links:
This search box allows for both general and specific querying and browsing of the database. Keywords used in the search can be general like 'DNA',
'RNA', 'hybrid' or specific like ligand names such as 'SPM' (3 letter code for the drug known as SPERMIN), full names such as 'Spermin' and
chemical formula like 'C10H26N4' (for SPM) and analogues.
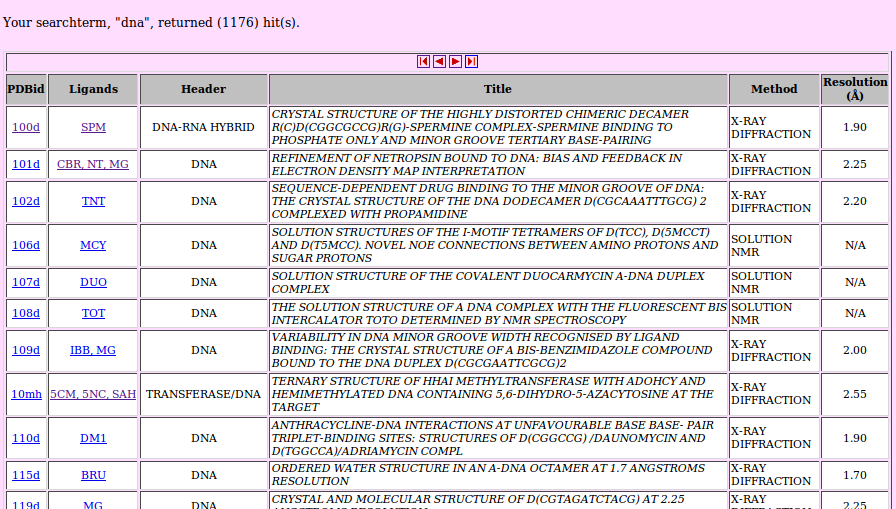
NALD displays a results list, Figure 3, which gives a summary list of known 3D structures in the PDB database for nucleic acid molecules (first column),
bound ligands (second column) and other useful information such as the title of the published molecules, method used to solve the structures and
Resolution at which the structures were solved.
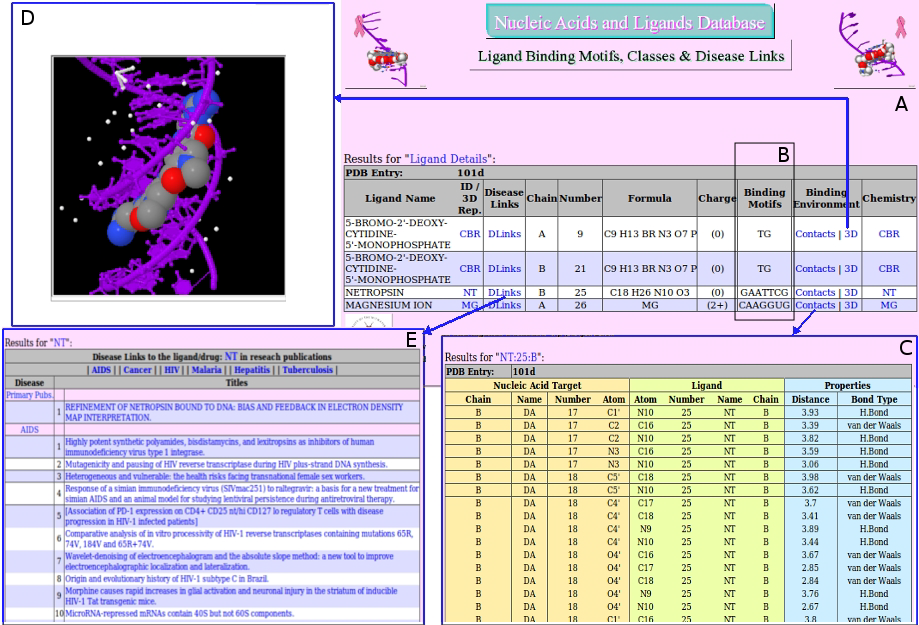
Detailed exploration of the ligand binding details with the NA molecule, calculated binding motif and disease links in addition to other useful
information about the ligand itself can be generated by clicking on ligand ids, second column, Figure 4. Further information about each PDB entry can
be displayed by clicking on the first column which retrieves the entry's summary from the LSE system.
Figure 4. Screenshot of result pages for ligands in the PDB entry 101d; DNA/Ligands
complex. A. overall list of ligands in the 104d. B. Calculated binding NA sequence
motifs for each ligand. C. Binding details with bond distances and possible types of
bonds between the ligand Netropsin (NT) and DNA. D Jmol 3D representation of the
DNA/NT binding. E. Disease links in relation with the ligand NT pointing to PubMed
abstracts.
Find Binding Motifs & Binding Classes:
NALD offers two types of binding motifs, the calculated binding motifs seen above (Figure 4B)
and empiric binding motifs reported in scientific literature
(e.g. Clark et.al., 1996 and Bunkenborg et.al., 2002) with detailed annotation of the motifs, binding sites, modes, classes and properties (Figure 5A).
Classes of NA–Ligands binding covers Intercalation, Modification, Addition and Cavity fitting in addition to binding modes of ligands to the DNA's
Minor Groove, Major Groove and both Minor/Major Grooves.
Detailed annotation is associated with each binding motif which includes the NA sequence, binding mode position(s) and types of NA bases involved
in the binding, Figure 5B. This is also reflected in the 3D representation, Figure 5C.
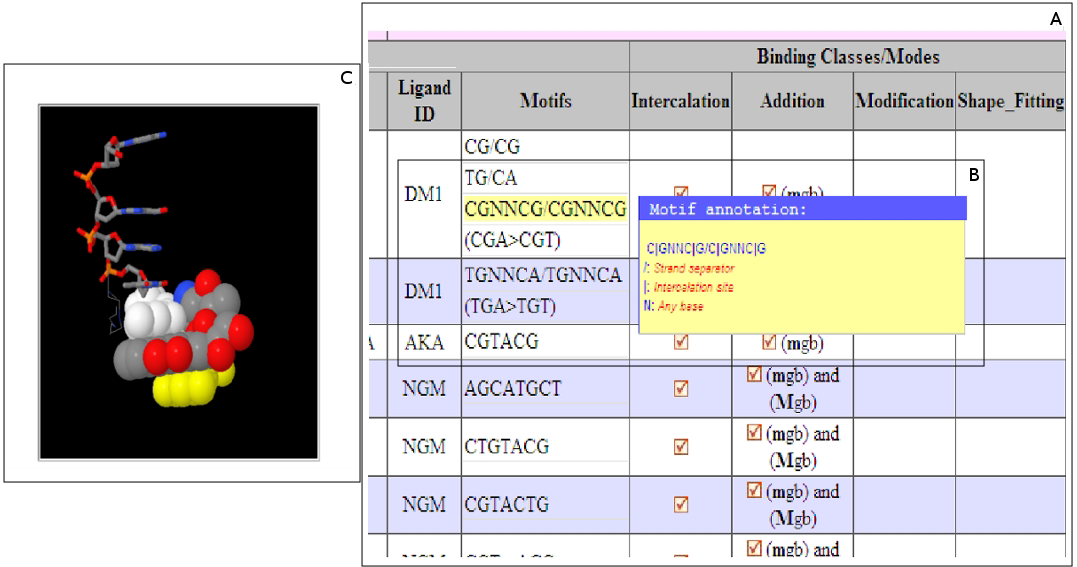
Figure 5. Screenshot of result pages for binding modes & motifs: A. Binding motifs (column Motifs) and binding modes (four columns under Binding Classes/Modes). B.
Detailed annotation of the binding motifs. This case shows an example of search output showing ligands, their specific binding motifs and classes/modes of binding.
The ligands DM shows two main modes of binding 1. Intercalation (| indicates which DNA bases binds the ligand in the motif) and 2. Addition which shows that some
ligands binds the DNA minor groove (mgb) or major groove (Mgb) or both at the same time. C. 3D representation showing, in this case, where exactly the ligand DM
binds in intercalation mode between DNA bases shown in white and yellow colours.
The search box also allows users to search for DNA, RNA or hybrid molecules that binding motifs containing particular bases such as 'CGC' or
searching for existing classes and modes of binding adopted by ligands such as typing 'Intercalation' or just 'I:' for finding those ligands that binds in
“Intercalation modes” and nucleic acids motifs binding them, and modification ('M:') for finding those ligands that causes “modification” when biding
nucleic acids.
Discussion
Nucleic acids are of great biological importance in all organisms and the inability to maintain integrity or tampering with nucleic acids leads to
disease. Small chemical molecules or ligands bind nucleic acids in various ways and thus therapeutic strategies are designed from the the
identification and study of binding details of these ligands including motifs bound to and modes of binding.
The NALD database, through the numbers of features described above, summarized in points below, has the potential to be instrumental in helping
with studies and processes involving the identification of potential nucleic acids targeted drugs and novel design of new drugs in the fight against
diseases currently thriving in the SADC region including the HIV/AIDS pandemic, cancer and other conditions such as tuberculosis (TB), malaria and
hepatitis.
NALD is a valuable resource for researchers in the fields of molecular biology, biochemistry,
and drug design. One of the main advantages of NALD is its user-friendly interface, which allows researchers to search for specific
interactions of interest using various criteria, such as the type of nucleic acid or ligand, the binding affinity, or the
three-dimensional structure of the complex. The database is also regularly updated with new information and features,
making it a valuable resource for researchers in the field.
Conclusion
NALD represents a significant contribution to the field of molecular biology and biochemistry, providing
a comprehensive resource for researchers interested in the interactions between nucleic acids and ligands. By providing detailed information on the
molecular interactions between these important biomolecules, NALD can aid in the development of new drugs and therapies.
In conclusion, NALD offers the following features important in biology in general and in relevant area of research including insights into
structure-function relationship and rational drug design considerations into fighting high profiled diseases:
1. NALD provides a point of acquisition for categorized information about nucleic acid binding ligand categories including binding motifs, types and
classes of the ligands and drugs.
2. The information contained and level of annotations provided in NALD paves the way to drug design strategies and identifying potential in various
ligands and drugs.
3. NALD supplies links to medically applicable information related to the ligands centered on current diseases facing large populations of SubSaharan
Africa.
4. NALD facilitates crossreferencing with other currently larger databases such as the PDB, UniProt and PubMed and provides a user friendly
environment for fast integrated data retrieval from a single system.
References
🕮 Rachedi, A., Madida, K. (2013). NALD: Nucleic Acids and Ligands Database. In: Amine, A., Otmane, A., Bellatreche, L. (eds) Modeling Approaches and Algorithms for Advanced Computer Applications. Studies in Computational Intelligence, vol 488. Springer, Cham. Available at: https://doi.org/10.1007/978-3-319-00560-7_36
🕮 Berman, H.M., Westbrook, J., Freng, Z., Gilliland., Bhat, T.N., Weissig, H., Shindyalov, I. N. and Bourne, P.E. (2000). The Protein Data Bank. Nucleic Acids Res., 28, 235242.
🕮 Bunkenborg,J., Behrens,C., Jacobsen,J.P. (2002). NMR Characterization of the DNA Binding Properties of a Novel Hoechst 33258 Analogue Peptide Building Block. Bioconjugate Chem., 13 (5), pp 927–936
🕮 Clark,G.R., Squire,C.J., Gray,E.J, Leupin,W., Neidle, S. (1996). Designer DNAbinding drugs: the crystal structure of a metahydroxy analogue of Hoechst 33258 bound to d(CGCGAATTCGCG)2. Nucleic Acids Res., 24(24), 4882–4889.
🕮 Dimitropoulos, D., Ionides, J. and Henrick K. (2006). Using PDBeChem to Search the PDB Ligand Dictionary. Current Protocols in Bioinformatics; Chapter 14:Unit14.3., 14.3.114.3.3
🕮 Sehnal, D., Bittrich, S., Deshpande, M., Svobodová, R., Berka, K., Bazgier, V., Velankar, S., Burley, S.K., Koča, J., Rose, A.S. (2021) Mol* Viewer: modern web app for 3D visualization and analysis of large biomolecular structures. Nucleic Acids Research. Available at: https://doi.org/10.1093/nar/gkab314
🕮 NALD. Nucleic Acids and Ligands Database. [Online] Available at: https://bioinformatics.univ-saida.dz/bit2/?arg=SB1
🕮 LSE: http://emboss.bioinf.wits.ac.za/lse
🕮 MySQL: The MySQL RDBMS. http://www.mysql.com
🕮 PMID: http://www.ncbi.nlm.nih.gov/PubMed