JSBB: Volume 2, Issue 2, August 2023 - STRUCTURE & FUNCTION ARTICLES
Research article: Master's research based.
Structural and Functional Binding Motifs in Porphyrin Proteins: Insights into Ligands and Biological Function
MEGHARBI Mohamed El-Mahdi, BITAR Mohamed and RACHEDI Abdelkrim📧
Laboratory of Biotoxicology, Pharmacognosy and biological valorisation of plants, Faculty of Sciences, Department of Biology, University of Saida - Dr Moulay Tahar, 20100 Saida, Algeria.
Published: 03 August 2023
Abstract
Understanding the underlying principles governing the relationship between protein structure and function is crucial for advancing our knowledge of biological functions, including those related to proteins and macromolecules in health and disease. Knowledge aspects generated in this work, in relation to protein ligand binding environment, would contribute in better understanding of biology mechanisms and is instrumental in the endeavours for designing novel drugs and developing biotechnology solutions.
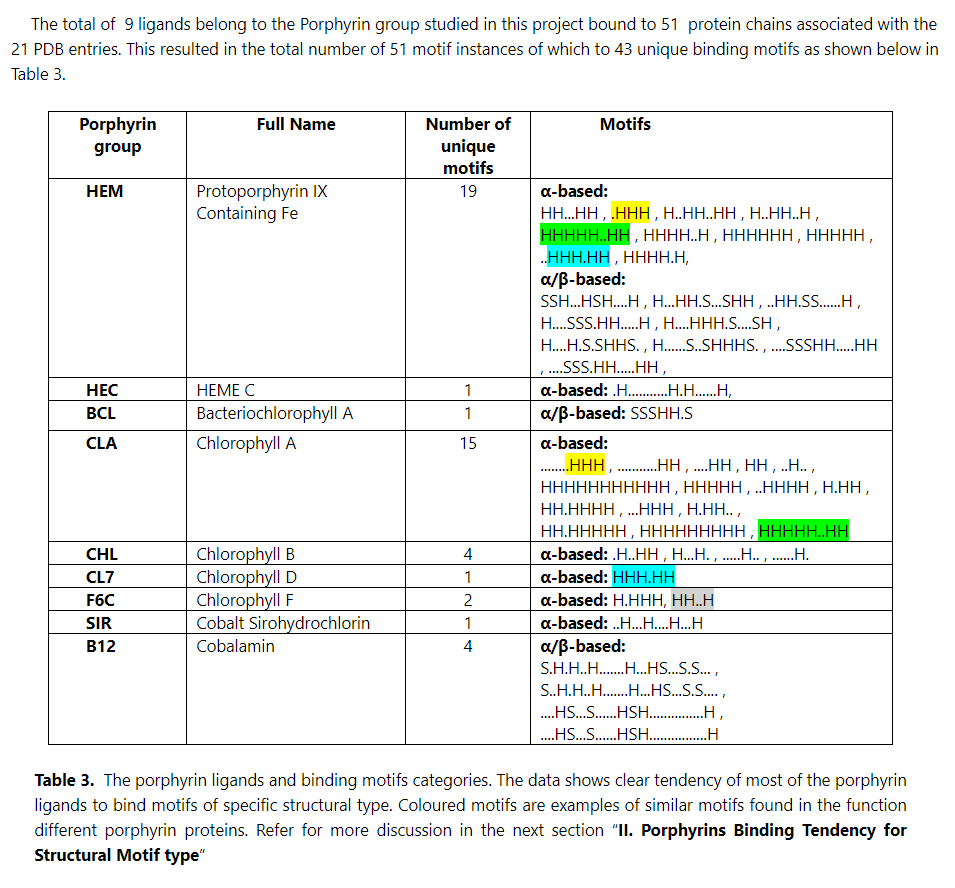
The research presented in this article examines a collection of 3D structures of porphyrin proteins, encompassing 51 full chains from 21 entries in the Protein Databank (PDB), originating from various organisms and species. The investigation identified, constructed and characterised 43 unique structural and functional binding motifs. These motifs are associated with essential biological functions such as oxygen transport, storage, light harvesting, and energy production.
The findings and analysis obtained in this work have been annotated in a database named Porphyrin Proteins Binding Structural Motifs (PPBSMs). This database has been made available online, through the web server of the University of Saida, to providing a valuable resource for scientists and researchers worldwide.
Availability:
The PPBSMs database is available freely here: https://bioinformatics.univ-saida.dz/prjs/ppbsms/
Key words
Porphyrin proteins, Porphyrin ligands, Heme, Chlorophyll, B12, Structural Bioinformatics, Structural & Functional Motifs, Ligand Binding Environment, Amino Acids, Residues, Databases.
Introduction
The biological function of proteins is intricately linked to their three-dimensional (3D) structure, which determines their biochemical activities. Within molecules, specific arrangements or patterns of atoms, functional groups, or molecular fragments, known as structural motifs, have significant implications for the physicochemical properties, biological activity, and pharmacokinetic behavior of therapeutic compounds.
Structural motifs are essential for protein function as they facilitate interactions with substrates, cofactors, and other ligands necessary for various biological processes. For instance, the helix-turn-helix motif (Aravind L, 2005) is a vital structural motif involved in DNA binding and gene transcription regulation, highlighting the complexity of identifying such motifs based solely on amino acid sequences.
The field of structural biology continually expands our knowledge of these motifs, and this project aims to explore their role in porphyrin containing proteins (Smith, K. M., & Ito, S., 2017, Wiley-VCH, 2011), such as Hemoglobin, Cytochromes, and other functionally important proteins. Porphyrin proteins are critical for essential processes in living organisms, including respiration and cellular metabolism. Dysregulation of these proteins can lead to diseases, including cancer. Therefore, characterizing the binding motifs associated with their functionality would greatly contribute to our understanding of their function and the identification of potential novel drugs.
Porphyrin proteins exhibit various structural motifs crucial for their specific functions. For example, hemoglobin and myoglobin, which are heme binding proteins (Trent JT, Hargrove MS, 2002) containing an Iron ion, possess a "globular" domain responsible for oxygen binding and transportation. Additionally, they have an "α-helical" domain that provides structural stability. Cytochromes (Ortiz de Montellano, P. R., 2005, Chiancone, E., & Ceci, P., 2010), on the other hand, feature a "cytochrome" motif involved in electron transfer reactions. Cobalamin or vitamin B12 (Scott, A. I., 1998), important for DNA synthesis and nerve function, relies on the porphyrin ring system that binds a Cobalt ion. Furthermore, porphyrin groups binding Magnesium ions are integral components of Chlorophyll A, B, D, and bacterial chlorophyll, essential parts of Photosystem I (Hill, R., & Bendall, D. S., 2014) and Photosystem II (Järvi, S., et. al., 2015) behind the photosynthesis and phosphorylation vital processes.
This research project aims to investigate and discover structural motifs associated with a range of proteins from different species that bind various types of porphyrin groups, including Heme and its derivatives, Chlorophyll types A, B, D, and F, and Cobalamin (Megherbi M. El.-M., Bitar M., 2023). By unraveling the relationship between these motifs and protein functionality, valuable insights can be gained, leading to potential advancements in drug discovery and a deeper understanding of pathological alterations in these proteins.
Materials and Methods
Results
Conclusion
This investigation research falls under the theme of Structural Bioinformatics and seeks to explore more the basis behind Structure-Function relationship in biological context of macromolecules; the proteins in the case of this study. Furthermore, the study draws attention to results that would touch upon distant evolutionary relations across species.
As revealed in the various analysis and deductions made in the Results and Discussions (Chapter III), this project has identified, defined and characterised a set of binding structural and functional motifs associated with a set of biologically important ligands/cofactors known collectively as Porphyrin groups. These ligands are relevant to vital biological function including oxygen transport, storage, light harvesting and energy production and more.
The project also identified the residues (amino acids) that are directly involved in the Porphyrin groups within the set of proteins selected in the study. The protein structural elements (α-helices and β-strands) and loop regions that compose the structural binding motifs are considered, by this study, as providing important physical support on which the actual functional elements, i.e. the residues, are mounted, with their individual and collective physical and chemical properties, to carry out the specific biological function of the porphyrin proteins.
The discovery of structural similarity between the porphyrin binding motifs across distant species and functions is indicative of evolutionary relation between these types of proteins that use similar chemical groups such a the porphyrin planar cycles to achieve different biological functions. This would open further venues of research and discovery in this field of study.
The definition of the ligand binding sites, i.e. the binding structural motifs, and construction of a database accessible online and that provide such important data and analysis to researchers in the field would be very useful in deeper analysis of the protein function in health and pathological cases, in studies related to phylogenetic analysis, 3D-structure predictions and rational drug design.
Future directions
The conclusions made above would be better confirmed and further explored using larger data sets of Porphyrin proteins and the ligand types used by them. Moreover, future work would include studies of the nature of the sequence of the motifs’ amino-acids and explore the effect of their consecutive arrangement in the binding of the porphyrin ligands.
References
🕮 Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM, Jul 2005, The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol Rev. 29(2):231-62. doi: 10.1016/j.femsre.2004.11.005.
🕮 Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., ... & Bourne, P. E., 2000, The Protein Data Bank. Nucleic acids research, 28(1), 235-242.
🕮 Chiancone, E., & Ceci, P., 2010, "The multifaceted capacity of cytochrome b5 in plants". Plant Signaling & Behavior, 5(1), 27-31.
🕮 Hill, R., & Bendall, D. S., 2014, Photosystem I: Structure and function. Essays in biochemistry, 56, 41–61. https://doi.org/10.1042/bse0560041).
🕮 Järvi, S., Suorsa, M., & Aro, E. M., 2015, Photosystem II repair in plant chloroplasts—Regulation, assisting proteins and shared components with photosystem II biogenesis. Biochimica et Biophysica Acta (BBA) - Bioenergetics, 1847(9), 900-909).
🕮 Megherbi M. El Mehdi, Bitar M., 2023, Bioinformatics based Characterization of Porphyrin Protein Binding Structural Motifs and Construction of a relevant Database: Unravelling some Essentials of Protein Structure-Function Relationships (https://bioinformatics.univ-saida.dz/prjs/ppbsms/). Master Thesis, Library of the University of Saida.
🕮 Nelson, N., & Yocum, C. F., 2006, Structure and function of photosystems I and II. Annual Review of Plant Biology, 57, 521-565.
🕮 Ortiz de Montellano, P. R., 2005, "Cytochrome P450: structure, mechanism, and biochemistry". Springer Science & Business Me.
🕮 Smith, K. M., & Ito, S. (Eds.), 2017, Porphyrins and related macrocycles: synthesis, spectroscopy, and potential applications. Royal Society of Chemistry.
🕮 Scott, A. I., 1998, Enzymatic synthesis of vitamin B12. Chemical reviews, 98(2), 491-516. doi: 10.1021/cr960426r.
🕮 Sayle, R. A., Milner-White E. J., 1995, RASMOL: biomolecular graphics for all, Trends Biochem Sci. Sep;20(9):374. doi: 10.1016/s0968-0004(00)89080-5.
🕮 Trent JT, Hargrove MS, 2002, "A ubiquitously expressed human hexacoordinate hemoglobin". The Journal of Biological Chemistry. 277 (20): 19538–19545.
🕮 Wiley-VCH, 2011, "Porphyrin". Encyclopedia of Inorganic and Bioinorganic Chemistry.