JSBB: Volume 2, Issue 2, August 2023 - STRUCTURE & FUNCTION ARTICLES
Concept article:
RBMSC, A Bioinformatics Tool for Generating Short Structural Motifs of Proteins and Nucleic Acids
RACHEDI Abdelkrim📧
Laboratory of Biotoxicology, Pharmacognosy and biological valorisation of plants, Faculty of Sciences, Department of Biology, University of Saida - Dr Moulay Tahar, 20100 Saida, Algeria.
Published: 15 August 2023
Abstract
In the realm of structural biology, understanding the conformational properties of biomolecules is of paramount importance. To this end, a innovative bioinformatics tool is introduced designed to create short structural motifs of proteins and nucleic acids in standard conformations. The folding of protein structures into specific functional units is characterised by local defined conformations mostly stretches in alpha helical and beta stands (extended) motifs. DNA structure though less complicated compared to proteins can exist in conformations, depending on water content, known as A, B and Z forms. This tool, named “Rapid Building of Protein/DNA Motifs in Standard 3D-Conformations” (RBMSC), offers a user-friendly interface that enables users to generate various short protein and DNA motifs. The motifs would enhance the study of local structural characteristics in biomolecules. By providing insights into the behaviour of specific amino acid and nucleic acid sequences, this tool contributes significantly to the field of molecular modelling and structural analysis.
Availability:
The RBMSC is available via the web address: https://bioinformatics.univ-saida.dz/bit2/?arg=MD1
Key words
Molecular Modelling, Standard conformations, Structural Motifs, Bioinformatics, Amino Acids, Nucleic Acids.
Introduction
Structural biology stands at the forefront of deciphering the intricate three-dimensional architectures that underlie the functionality of biomolecules. Within this discipline, a pivotal aspect involves understanding the conformational properties of proteins and nucleic acids, as these molecular structures inherently dictate their biological roles. To address this fundamental need, we present an innovative bioinformatics tool tailored to generate short structural motifs of proteins and nucleic acids in well-defined, standard conformations. This tool, named "Rapid Building of Protein/DNA Motifs in Standard 3D-Conformations" (RBMSC), serves as a transformative resource, enabling researchers to delve into the structural intricacies of biomolecules at the local level.
Proteins, being central to cellular processes, intricately fold into functional units characterized by local defined conformations, often encompassing stretches of alpha helices and extended beta strands [1]. Understanding the spatial arrangement of these structural motifs is pivotal in unraveling protein functions and interactions. The accurate depiction of these motifs aids researchers in deciphering the relationship between sequence and structure, thus guiding investigations into molecular behavior [2].
In parallel, the structural configuration of DNA, while comparatively less complex than proteins, exhibits distinct conformational variations influenced by environmental factors, particularly water content. DNA can adopt conformations known as A, B, and Z forms [3]. The ability to elucidate the specific conformational preferences of DNA sequences contributes profoundly to unraveling the molecular basis of DNA-protein interactions, DNA packaging, and other DNA-dependent processes.
The RBMSC tool, which we present here, is a response to the need for a user-friendly interface that bridges the gap between researchers and the exploration of biomolecular structural motifs. By generating short protein motifs and DNA conformations, the tool facilitates the investigation of local structural characteristics. Users can uncover the behaviors and preferences of specific amino acid and nucleic acid sequences, advancing the field of molecular modeling and structural analysis.
In the subsequent sections of this article, we delve into the intricacies of the RBMSC tool, detailing its methodology, user interface, applications, and future developments. Through this endeavor, we aim to empower researchers and students alike with an invaluable resource that unlocks the local structural nuances of biomolecules.
Materials and Methods
Methodology:
The core functionality of our tool lies in its ability to generate short structural motifs in well-established conformations. The protein motifs include right-handed α-helices, left-handed α-helices, 3-10 helices, π-helices, parallel β-strands, anti-parallel β-strands, β-turn type I, and β-turn type II. In addition, combinations of the basic motifs into constructs like α-helix_β-turn_α-helix motifs, also found in proteins, can be generated by the tool. Similarly, the DNA motifs can be in DNA A-form and DNA B-form. The tool achieves this through an intuitive interface that allows users to select specific amino acids or nucleic acids from relevant clickable lists.
User Interface:
The user-friendly interface is designed to accommodate both novice and experienced users, Figure 1. Users can input short amino acid or nucleic acid sequences by mouse selection from comprehensive lists of relevant building blocks, Figure 1.(A3) and (B3). These sequences are then transformed into the selected structural motif, allowing users to visualize and study the local conformational preferences of the biomolecules, see next section.
Figure 1. RBMSC interface. A for creating protein motifs where users can select simple motifs A1 or combined motifs A2. B to be selected for generating DNA motifs A or B forms B1 in Single or Double strand B2. Users can build their desired sequence for the motif by picking Amino Acids A3 or Nucleic Acids B3 and sequence shows up in the text-boxes A4 or B4. Users then click the button “Build 3D-Structure” which does the work and display the motif structures as shown in Figure 2 and 3.
Data Source:
The creation of reliable structural motifs necessitates the utilisation of well-established geometry parameters including bond-distances, bond-angles, and torsion angles. To achieve this, we leveraged data from the Protein Data Bank (PDB) [4] and pertinent scientific literature [5]. High-resolution structures available in the PDB serve as a primary source of reference for mean standard values of these structural parameters.
Structural Parameter Retrieval:
Bond distances, bond angles, and torsion angles were extracted from a curated selection of high-resolution protein and nucleic acid structures in the PDB. The chosen structures span a wide range of biomolecules and conformations, ensuring the representation of diverse molecular contexts. These structural parameters were then analysed to determine mean standard values for each motif conformation [6, 7].
Motif Generation:
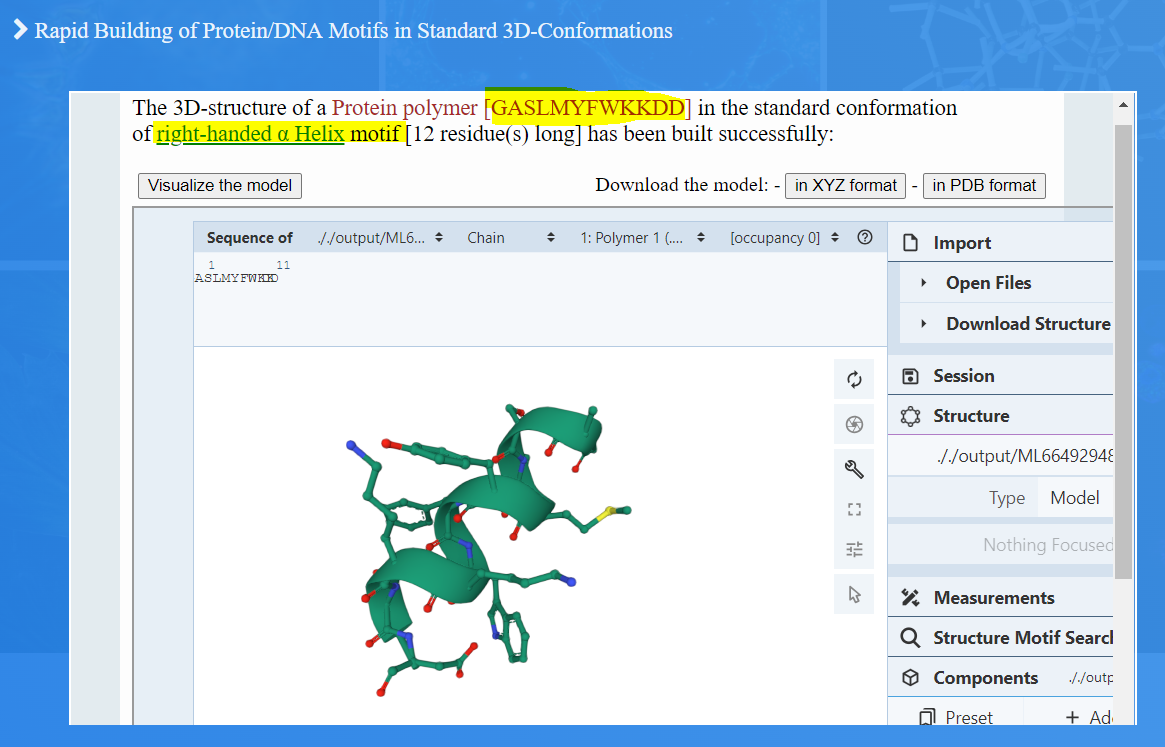
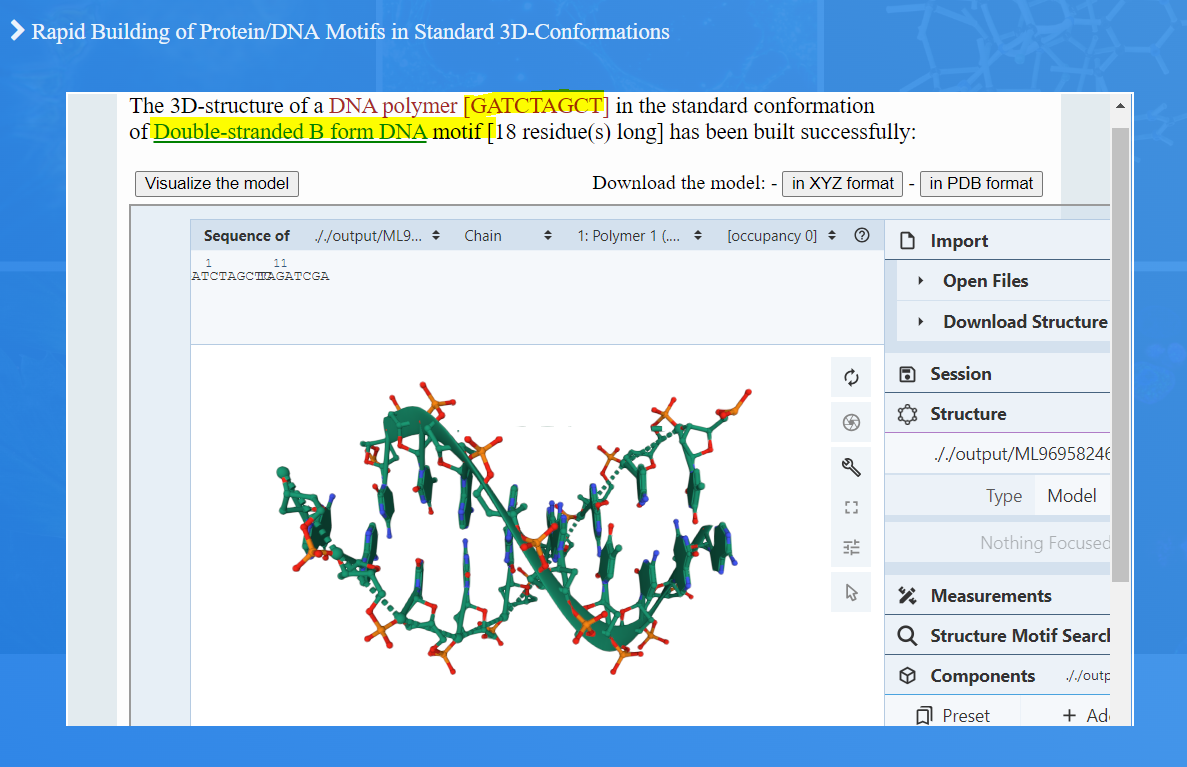
Upon establishing the mean standard values for bond distances, bond angles, and torsion angles, the RBMSC tool employs the data to generate three-dimensional representations of the selected motifs. When users input a specific sequence and motif type, Figure 1.(A4) & (B4), the tool outputs a coherent and realistic 3D-representation of the chosen motif, Figures 2 and 3.
Figure 2. RBMSC display of the user desired motif in 3D view. In this case, the motif selected by the user in Figure 1 is displayed in cartoon display together with side-chains of the Amino acids (sequence) in ball & Stick presentation. Using the mouse, users can rotate, zoom out or in the motif and do other graphics manipulations. Users can also download the coordinates of the motif structure from the buttons provided at the top of the display window.
Figure 3. An RBMSC example for generating the structure of double-stranded DNA B-form motif of a user selected sequence.
Conformational Preferences:
Validation, Accuracy and Limitations:
To ensure the accuracy and fidelity of the generated motifs, we conducted validation tests against a set of benchmark structures with known conformations. The motifs produced by RBMSC were compared to the reference structures, and the root mean square deviations (RMSD) were acceptably low. However, a more comprehensive comparison of the generated motifs with larger sets of structural data from the PDB is planned in future versions to demonstrate that RBMSC accurately reproduces the expected conformations within acceptable RMSD thresholds [8].
While the RBMSC tool strives to provide accurate representations of structural motifs, it is important to acknowledge certain limitations. The motifs are not energy minimised and the use of mean standard values may not fully capture the inherent flexibility and variation that biomolecules can exhibit. Additionally, the tool's accuracy is contingent upon the quality and diversity of the input data from the PDB.
Future Enhancements:
As part of our ongoing efforts, we are dedicated to refining the accuracy of motif generation by continually updating our dataset with the latest high-resolution structures from the PDB. Additionally, we are exploring the incorporation of machine learning techniques to enhance the tool's predictive capabilities and accommodate variations in structural parameters.
Conclusion
In conclusion, our bioinformatics tool presents a novel approach to generating short structural motifs of proteins and nucleic acids in standard conformations. By providing a user-friendly interface and diverse motif options, RBMSC empowers researchers and students to explore local conformational preferences in biomolecules in addition to individual amino acids and nucleic acids. This resource would contribute to the advancement of structural biology studies and paves the way for deeper insights into the relationship between sequence and structure in proteins and DNA.
References
🕮 1. Dill, K. A., & MacCallum, J. L. (2012). The protein-folding problem, 50 years on. Science, 338(6110), 1042-1046.
🕮 2. Ramachandran, G. N., Ramakrishnan, C., & Sasisekharan, V. (1963). Stereochemistry of polypeptide chain configurations. Journal of Molecular Biology, 7(1), 95-99.
🕮 3. Saenger, W. (1984). Principles of nucleic acid structure (Vol. 1). Springer Science & Business Media.
🕮 4. Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., ... & Bourne, P. E. (2000). The Protein Data Bank. Nucleic Acids Research, 28(1), 235-242.
🕮 5. Cui, X., Li, S.C., Bu, D., Alipanahi, B. & Li, M. (2013). Protein Structure Idealization: How accurately is it possible to model protein structures with dihedral angles?. Algorithms Mol Biol 8, 5. https://doi.org/10.1186/1748-7188-8-5
🕮 6. Engh, R. A., & Huber, R. (1991). Accurate bond and angle parameters for X-ray protein structure refinement. Acta Crystallographica Section A: Foundations of Crystallography, 47(4), 392-400.
🕮 7. Karplus, P. A., & Schulz, G. E. (1985). Prediction of Chain Flexibility in Proteins: A Tool for the Selection of Peptide Antigens. Naturwissenschaften, 72(4), 212-213.
🕮 8. Kleywegt, G. J., & Jones, T. A. (1997). Detecting folding motifs and similarities in protein structures. Methods in enzymology, 277, 525-545.