JSBB: Volume 1, Issue 4, February 2023 - BIO-DATABASES
Concept article: Minireviews
Bioinformatics: An Exciting Field of
Science - Importance and Applications
RACHEDI Abdelkrim
Laboratory of Biotoxicology, Pharmacognosy and biological valorisation of plants, Faculty of Sciences, Department of Biology, University of Saida - Dr Moulay Tahar, 20100 Saida, Algeria.
📧 E. mail: abdelkrim.rachedi@univ-saida.dz
Published: 01 February 2023
Abstract
Bioinformatics is a rapidly growing field that combines biology, computer science, and information technology to analyze and interpret biological data. In this article, we will explore the basics of Bioinformatics and delve into the databases that play a crucial role in the field. The article will include an overview of UniProt, Genbank, and PDB and demonstrate how these databases can be used in research and real-world applications.
Under the theme of usage examples of Bioinformatics, insights are also given in relation to the implementation of these databases and dedicated software such as the Modeller, AlphaFold2 and RoseTTAFold in the area of predicting protein structure and function and exploration of disease mechanisms.
Key words
Biological Data, Analysis, Databases, Uniprot, Genbank, PDB, Structure Prediction, AI Based Structure Modelling.
Introduction
Bioinformatics is a relatively new discipline that emerged with the explosion of genomic data. The field aims to extract meaningful information from large amounts of biological data and to apply computational and statistical methods to better understand biological systems. Over the years, this field has become an essential tool for scientists and researchers, providing insights into various areas of biology, including genetics, evolution, and disease.
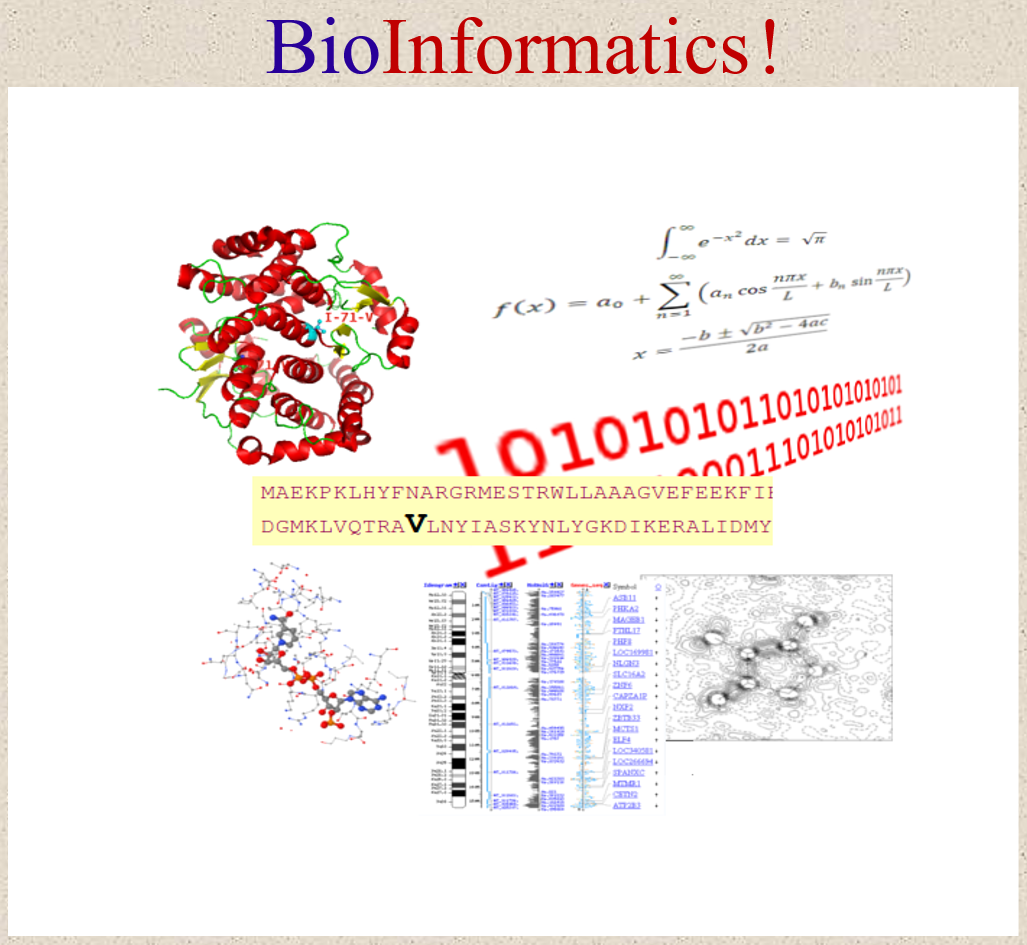
In the context of the scientific fields paradigm, bioinformatics should be seen an interdisciplinary field that combines computer science, statistics, and biology to analyze and interpret biological data, Figure 1. This domain emerged as a field in academia and research in the late 20th century as the amount of biological data generated through genomics and other high-throughput technologies rapidly increased.
Today, Bioinformatics is a critical component of modern biological research, providing the tools and techniques necessary to store, analyze, and interpret the vast amounts of data generated by high-throughput experiments. Research students interested in pursuing a career in Bioinformatics will need to have a strong background in computer science, biology, and statistics, as well as a deep understanding of the databases and tools used in the field.
Figure 1. Illustation of Bioinformatics as an interdisciplinary field that combines computer science, statistics, and biology
Methods and Databases
One of the key components of Bioinformatics is the use of databases. These databases store and provide access to large amounts of biological information, such as gene sequences, protein structures, and metabolic pathways. Three of the most widely used databases in Bioinformatics are UniProt, Genbank, and PDB.
UniProt:
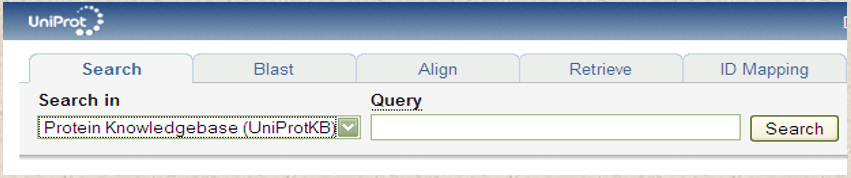
UniProt is a comprehensive, publicly available, and freely accessible database of protein information, Figure 2. It provides information on over 200 million protein sequences, including functional and taxonomic information, as well as protein-protein interactions. UniProt is an excellent resource for researchers, allowing them to search for specific proteins and to analyze their properties, functions, and interactions.
Figure 2. Partial screenshot shows the interface search bar of the UniProt database.
Available here: https://www.uniprot.org/
Available here: https://www.uniprot.org/
Genbank:
Genbank is a database of DNA and RNA sequences, including both coding and non-coding sequences. It currently provides information on over 19.6 trillion base pairs from over 2.9 billion nucleotide sequences and serves as a critical resource for genome sequencing projects and comparative genomics research. In addition to raw sequence data, Genbank also provides annotated information, such as gene names, functional descriptions, and taxonomic information.
Figure 3. Screenshot shows part the interface page of the Genbank database.
Available here: https://www.ncbi.nlm.nih.gov/genbank/
Available here: https://www.ncbi.nlm.nih.gov/genbank/
PDB:
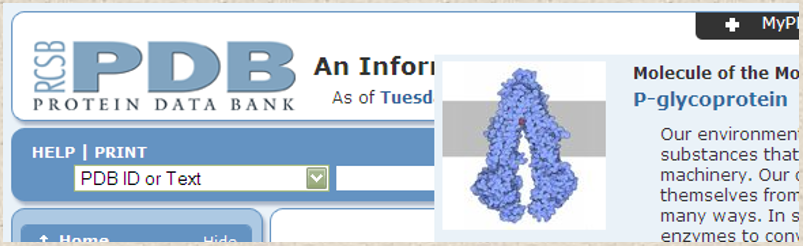
The Protein Data Bank (PDB) is a database of protein structures, including X-ray crystallographic and NMR structures. Todate, in 2023, it provides access to over 200,700 protein structures and serves as a valuable resource for researchers studying protein function and interaction. PDB provides information on protein structures, including 3D models and atomic coordinates, as well as functional annotations and information on protein-protein interactions.
Figure 4. Partial screenshot shows the interface search bar of the Protein databank - PDB.
Available here: https://www.rcsb.org/
Available here: https://www.rcsb.org/
Usage Examples
To demonstrate the practical applications of these databases, we will provide a few usage examples.
UniProt to study protein interactions
A researcher is studying the interactions between two specific proteins in a cellular pathway. They use UniProt to search for the sequences of these proteins and then use the database to identify potential interactions with other proteins. By analyzing the data available in UniProt, the researcher can gain insights into the functional relationships between these proteins and their role in the cellular pathway.
Genbank to compare genomes
A scientist is studying the evolution of a particular species and wants to compare its genome with that of related species. They use Genbank to access the DNA sequences of these species and perform a comparative analysis to identify similarities and differences between the genomes. By analyzing the data in Genbank, the scientist can gain insights into the evolution of the species and its relationships to other species.
PDB to study protein structure
A researcher is studying the structure of a specific protein and its interactions with other proteins. They use PDB to search for the 3D structure of the protein and analyze its atomic coordinates, functional annotations, and information on protein-protein interactions. By analyzing the data in PDB, the researcher can gain insights into the structure of the protein under study and potentially infer its functional properties.
Predicting Protein Structure and Function
Protein structure and function are two crucial aspects of bioinformatics research. Protein structure prediction is essential for understanding how proteins interact with each other and with other biomolecules in a cell. The Protein Data Bank (PDB) is a crucial resource for this research, as it provides detailed information on the three-dimensional structure of proteins.
One common approach to predicting protein structure is comparative modeling, which involves aligning the amino acid sequence of a target protein with the structure of a related protein with known structure. Researchers can use the UniProt database to identify related proteins and the PDB to access their structures.
Another approach to protein structure prediction is de novo modeling, which involves building a structure from scratch based on the protein's amino acid sequence. In this case, researchers can use tools such as the template-based methods ROSETTA and MODELLER, which are available through the PDB, to generate models of the protein structure.
Protein structure prediction has been greatly improved both in methodology and accuracy. Artificial Intelligence (AI) based methods represented by AlphaFold2 and RoseTTAFold have successfully been used in producing outstandingly accurate non-homology-based models compared to the classical methods when check against experimentally determined structures available in the PDB.
The high accuracy of the models created by these AI based methods has the potential to greatly aid in drug discovery, as well as improve our understanding of how proteins function and interact with each other. The development of these models marks a major step forward in the field of protein folding prediction, and it will be interesting to see how they continue to evolve and impact the field in the coming years.
Predicting protein function is also a critical aspect of bioinformatics research. In this case, researchers can use the UniProt database to identify proteins with similar functions and the PDB to access information on their structures. By comparing the structures of proteins with similar functions, researchers can gain insights into the relationships between structure and function.
Additionally, researchers can use functional annotation tools, such as InterPro and Gene Ontology, to predict the function of a protein based on its sequence. These tools are available through the UniProt database and can be used to analyze large amounts of data in a high-throughput manner.
Understanding Disease Mechanisms
Understanding the molecular mechanisms underlying diseases is a crucial aspect of bioinformatics research. The UniProt and Genbank databases provide information on the genomic sequence and functional annotations of disease-causing genes and proteins.
For example, researchers can use the UniProt database to identify mutations in disease-causing genes that result in the production of altered or non-functional proteins. By analyzing the structures of these proteins, researchers can gain insights into the molecular mechanisms underlying the disease.
In addition, researchers can use the Genbank database to identify genetic variations associated with diseases, such as single nucleotide polymorphisms (SNPs) and copy number variations (CNVs). By analyzing these variations, researchers can gain insights into the genetic basis of diseases and develop new diagnostic and therapeutic approaches.
Conclusion
Bioinformatics is a rapidly evolving field that has revolutionized the way researchers do thier studies and plays a critical role in modern biology.
The UniProt, Genbank, and PDB databases are essential resources for bioinformatics research, providing access to large amounts of data on genomic sequences, protein structures, and functional annotations.
In this article, we have discussed the importance of bioinformatics for graduate and post-graduate students and provided examples of how these databases can be used in research. We have also emphasized the importance of comparative modeling, de novo modeling, functional annotation, and disease mechanisms in bioinformatics research.
The field of bioinformatics is constantly expanding, and new developments in technology and computational approaches will continue to drive its growth. Institutions and individuals interested in this field must have a strong foundation in both biology and computer science and be familiar with the use of databases and computational tools.
References
🕮 A. Sali, T. Blundell, "Modeller: Generation and Refinement of Homology-Based Protein Models" in Methods in Enzymology, vol. 374, pp. 461-491, (2003).
🕮 A. Sali, "Modeller: Protein Structure and Function Predictions" in Current Protocols in Bioinformatics, vol. 49, (2017).
🕮 Altschul, S. F., Gish, W., Miller, W., Myers, E. W., & Lipman, D. J. (1990). Basic local alignment search tool. Journal of molecular biology, 215(3), 403-410.
🕮 Bryne, J. C., Goodsell, D., & Sansom, M. S. (2005). Structure of ligand-free and liganded protein kinases. Nature, 435(7044), 1162-1169.
🕮 Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., & Madden, T. L. (2009). BLAST+: architecture and applications. BMC bioinformatics, 10(1), 421.
🕮 Davis, S., & Bougueleret, L. (2019). Bioinformatics and genomic data analysis. In From DNA to RNA (pp. 181-202). Humana, New York, NY.
🕮 GenBank. (2023). NCBI. https://www.ncbi.nlm.nih.gov/genbank/
🕮 Kanehisa, M., & Goto, S. (2000). KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic acids research, 28(1), 27-30.
🕮 PDB. (2023). RCSB Protein Data Bank. https://www.rcsb.org/
🕮 S.M. Kim, et al., "Improved protein structure prediction using potentials from deep learning" in Nature, vol. 574, pp. 84-90, (2019).
🕮 R. Zhao, et al., "RoseTTAFold: An Improved AlphaFold-Based Method for Protein Structure Prediction" in bioRxiv, (2022).
🕮 UniProt. (2023). EBI-UniProt. https://www.uniprot.org/
🕮 UniProt Consortium. (2023). UniProt: the Universal Protein Resource. Nucleic acids research, 51, D523-D531.
🕮 Zhang, J., Liu, X., Xie, X., & Sun, F. (2005). CD-HIT: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics, 21(13), 1658-1659.