Protein structure knowledge can help fight bacterial food poisoning.
Abdelkrim RACHEDI
Laboratory of Biotoxicology, Pharmacognosy and biological valorisation of plants, Faculty of Sciences, Department of Biology, University of Saida - Dr Tahar Moulay, 20100 Saida, Algeria.
📧 E. mail: abdelkrim.rachedi@univ-saida.dz
Published: 17 January 2023
Bile is a component essential for intestinal absorbtion of fats in the Human and animals. Some bacterial pathogens that can thrive in the gut envionement are equipped with sensery systems for bile that enable them to know the nature of the environment suitable for growth and launching severe infections.
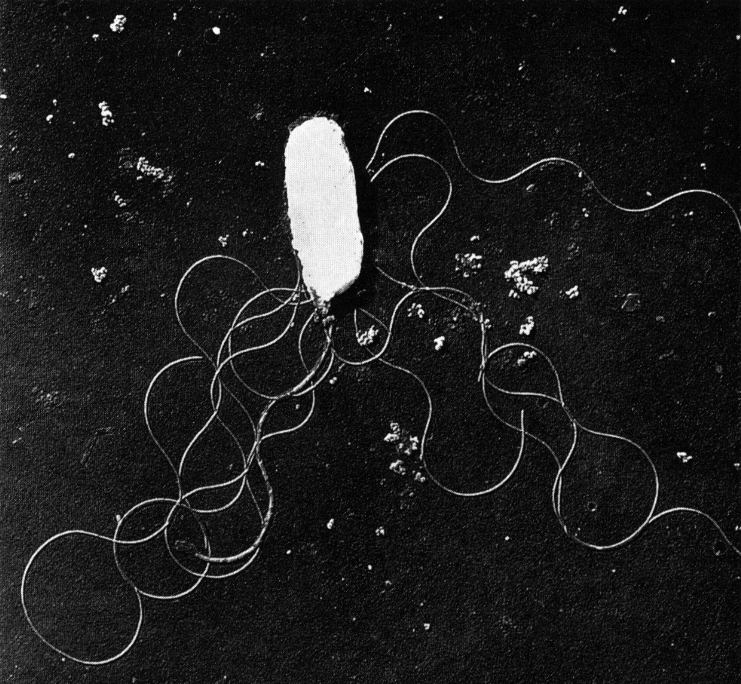
The bacterium Vibrio parahaemolyticus is poisinous species and can cause gastrointestinal illness and acute gastroenteritis. This happens when some types of oysters and similar shellfish are eaten undercooked or halfdone, Figure 1. (Yabuuchi E. et al., 1974).
This bacteria depend, in their infection of the digestive system and the release of toxins (in the intestine), on a number of systems, including a sensor system consisting of a complex of two transcription factors (proteins), VtrA/VtrC complex (Gotoh K. et al., 2010). They act as a sensor for bile salts / acids in the intestine, where it informs the bacteria that it is, now, present in the digestive system. This activates the pathogenic type III secretion system responsible for liberating toxins in the intestine leading to cases of food poisoning and to the accompanying pathological symptoms.
Figure 1. Electron micrograph of V. parahaemolyticus WP-1 on MMOF agar, 20 C 16 hr, palladium shadowed, •~ 25 000. Note polar flagellum is thicker than peritrichous flagella.
Customized from “Flagellar morphology of Vibrio parahaemolyticus (Fujino et al) Sakazaki, Iwanami and Fukumi 1963“ (Yabuuchi E. et al. 1974)
Customized from “Flagellar morphology of Vibrio parahaemolyticus (Fujino et al) Sakazaki, Iwanami and Fukumi 1963“ (Yabuuchi E. et al. 1974)
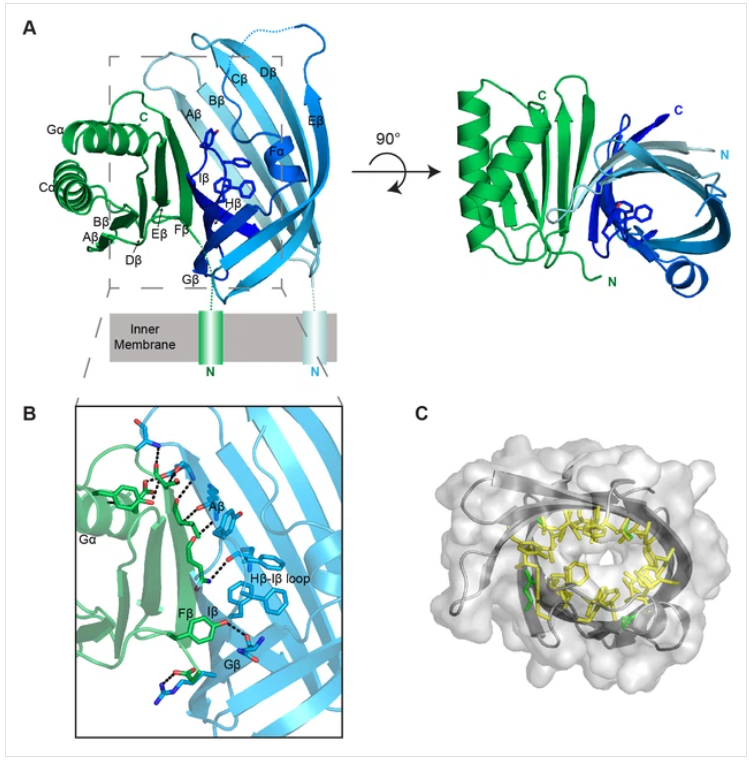
The 3D-structures of the periplasmic domains of the VtrA/VtrC heterodimer, in Figure 2, with1️⃣ and without2️⃣ bile saltreveals a β-barrel with a hydrophobic inner cavity, Figures 2(A) and (C). Additionally, biophysical and mutational analysis demonstrated that the hydrophobic cavity binds bile salts and activates the virulence cascade.
Figure 2. (A) Cartoon representation of the periplasmic domain complex formed by VtrA (green) and VtrC (blue, light to dark gradient from N-terminus to C-terminus). Side chains of Hβ-Iβ loop residues are shown as sticks. (B) Detailed view of the VtrA/VtrC interface. Selected residues that form polar contacts (black dashed lines), as well as potential bile salt binding residues are shown as sticks. (C) Overlay of surface and ribbon models of VtrC showing interior cavity. Side chains of residues lining the cavity are shown as sticks in yellow for hydrophobic residues (Ala, Val, Ile, Leu, Met, Phe, Tyr, Trp) and green for all other.
Depicted from “Bile salt receptor complex activates a pathogenic type III secretion system“ (Li P. et at., 2016)
Depicted from “Bile salt receptor complex activates a pathogenic type III secretion system“ (Li P. et at., 2016)
Structural study of this protein complex and subsequent analysis showed exactly how bile salts/acids such as taurodeoxycholate bind with these bacterial enzymes (Li P. et at., 2016).
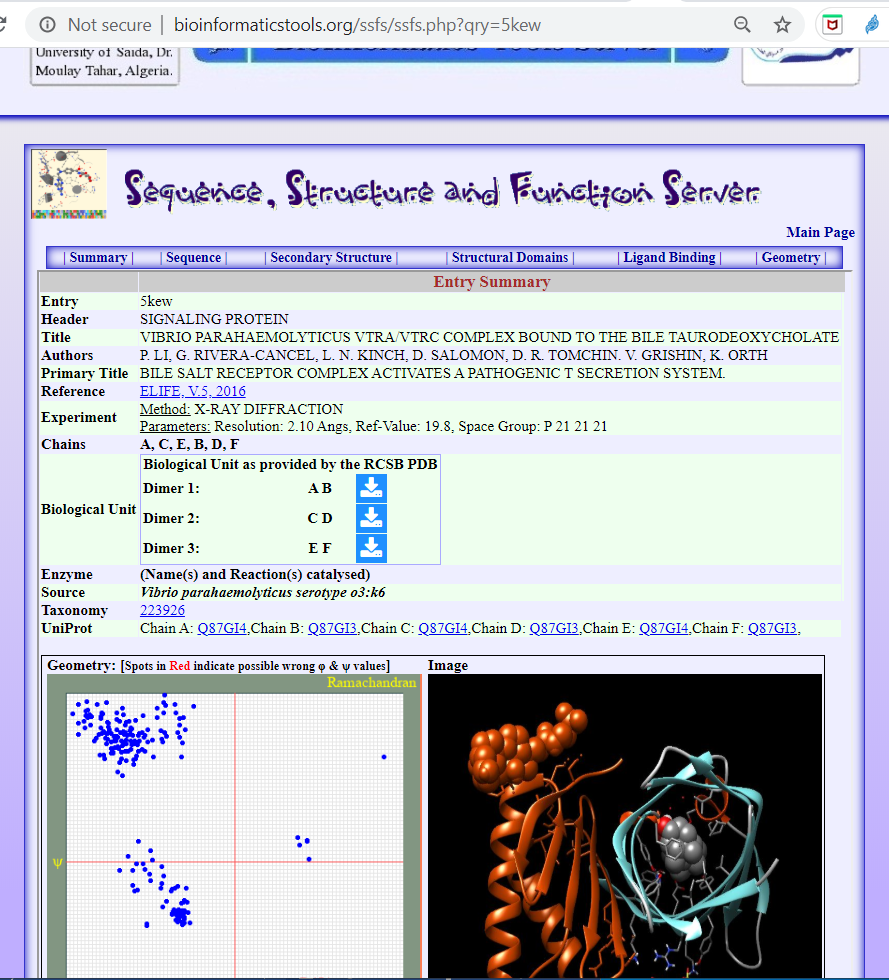
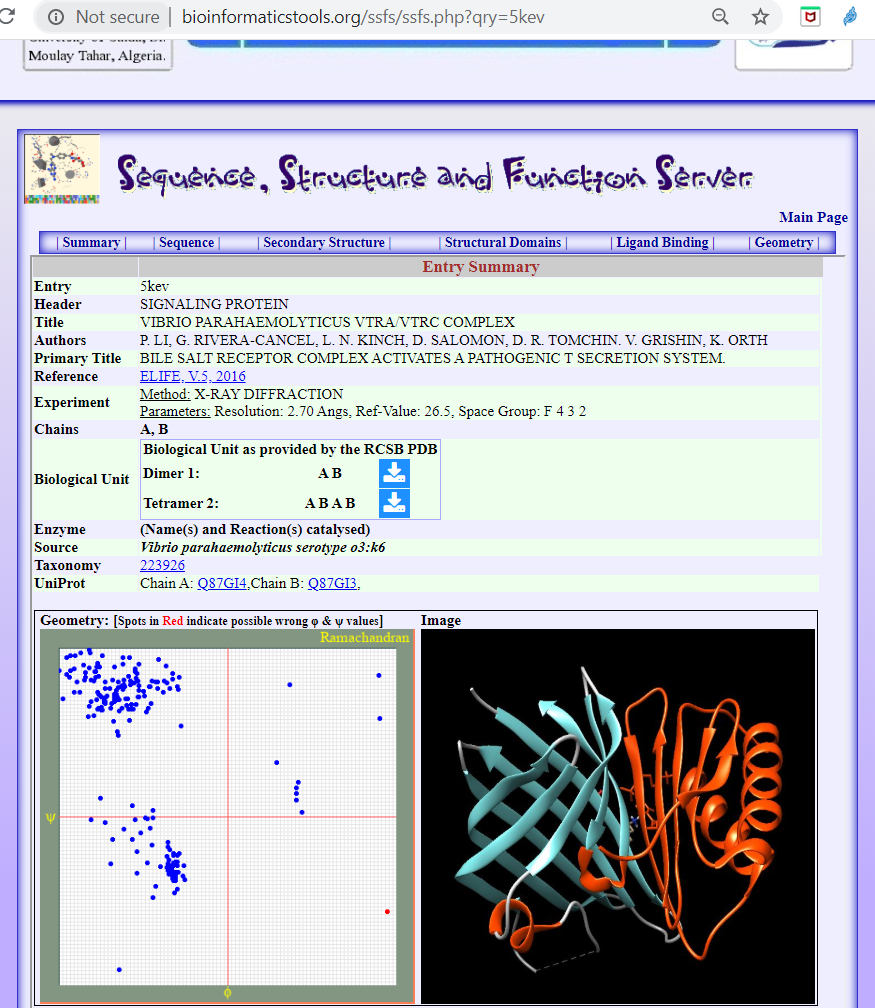
The structures are found annotated in the Protein Databank (PDB) denoted with codes 5kew for the VtrA/VtrC dimer in complex with the bile salt taurodeoxycholate, Figure 3., and the PDB code 5kev for the dimer in apo-form (without ligand), Figure 4.
These can be explored using the application SSFS⭕ by the Bioinformatics Server, Department of Biology, Saida University, Algeria, (Golovin A. et. al. 2005 - related reference), links below:
These can be explored using the application SSFS⭕ by the Bioinformatics Server, Department of Biology, Saida University, Algeria, (Golovin A. et. al. 2005 - related reference), links below:
These structures can also be explored from the Protein Data Bank Europe (PDBe) at:
Figure 3. The structure of the Vibrio Parahaemolyticus Vtra/Vtrc Complex Bound To The Bile Taurodeoxycholate Click menu “Ligand Binding“ for finding details of the binding between VtrA/VtrC heterodimer and the ligand Taurodeoxycholate (bile salt).
See: https://bioinformaticstools.org/ssfs/ssfs.php?qry=5kew
See: https://bioinformaticstools.org/ssfs/ssfs.php?qry=5kew
Figure 4. The structure of the Vibrio Parahaemolyticus Vtra/Vtrc Complex
See: https://bioinformaticstools.org/ssfs/ssfs.php?qry=5kev
See: https://bioinformaticstools.org/ssfs/ssfs.php?qry=5kev
It is worth mentioning that these studies along with details such as the binding of VtrA/VtrC heterodimer to the bile🔶 can contribute into the manufacturing of novel drugs using the Rational Drug Design method that can lead to deceiving such enzymes and competatively bind with them so they are unable to sense bile salts/acids, which thwarts the process of releasing toxins in the intestine, and thus obtaining protection from food poisoning.
Follow the link below for details about the molecular mechanism behind this bacterial system, (Li P. et at., 2016):
🔶Details of the binding enviroment is provided by the SSFS tool as shown in Figure 3 by clicking the menu button "Ligand Binding"
⭕ SSFS: Sequence, Structure and Function Server by the Structural Biology & Bioinformatics Groups, Biology Dept, Faculty of Science, University of Saida, Algeria
نظام: التركيب الأولي، التركيب الفراغي و الوظيفة البيولوجية ، فريق البيولوجيا ثلاثية
الأبعاد و المعلوماتية الحيوية، قسم البيولوجيا، كلية العلوم، جامعة سعيدة، الجزائر
References
E Yabuuchi, T Miwatani, Y Takeda, M Arita " Flagellar morphology of Vibrio parahaemolyticus (Fujino et al) Sakazaki, Iwanami and Fukumi 1963 " Jpn J Microbiol. 1974 Jul;18(4):295-305.
- in PubMed
Kazuyoshi Gotoh, Toshio Kodama, Hirotaka Hiyoshi, Kaori Izutsu, Kwon-Sam Park, Rikard Dryselius, Yukihiro Akeda, Takeshi Honda, Tetsuya Iida " Bile acid-induced virulence gene expression of Vibrio parahaemolyticus reveals a novel therapeutic potential for bile acid sequestrants " PLoS One. 2010 Oct 13;5(10):e13365.
- in PubMed
Peng Li, Giomar Rivera-Cancel, Lisa N Kinch, Dor Salomon, Diana R Tomchick, Nick V Grishin, Kim Orth "Bile salt receptor complex activates a pathogenic type III secretion system" Elife. 2016 Jul 5;5:e15718.
- in PubMed
Golovin A., Dimitropoulos D., Oldfield T., Rachedi A. and Henrick "MSDsite: A Database Search and Retrieval System for the Analysis and Viewing of Bound Ligands and Active Sites." PROTEINS: Structure, Function, and Bioinformatics 58(1): 190-9 2005
- in PubMed