JSBB: Volume 2, Issue 4, January 2024 - ANTIBIOTICS RESISTANCE ARTICLES
Research article: Master's research based.
🕮 Al Aukidy, M., Al Chalabi, S., & Verlicchi, P. (2018). Hospital wastewater treatments adopted in Asia, Africa, and Australia. Hospital wastewaters: characteristics, management, treatment and environmental risks, 171-188.
🕮 Baumont, S. (2004). Réutilisation des eaux usées épurées : risques et faisabilité en Ile de France. Toulouse.
🕮 Bradford, P. A. (2001). Extended-spectrum β-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clinical microbiology reviews, 14(1), 933-951.
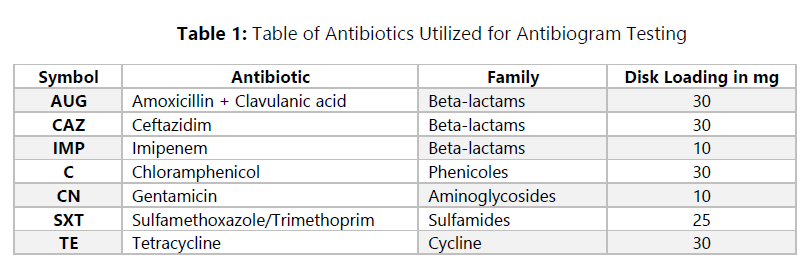
🕮 CLSI. (2012). Performance Standards for Antimicrobial Susceptibility Testing. Clinical and Laboratory Standards Institute. 26th Edition, M100S, Wayne. USA.
🕮 Cockerill, F. R. (2012). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow. Approved Standard—Ninth Edition. CLSI, 12.
🕮 Ekhaise, F., & Omavwoya, B. P. (2008). Influence of hospital wastewater discharged from University of Benin Teaching Hospital (UBTH), Benin City on its receiving environment. American-Eurasian J Agric Environ Sci,, 4(4), 484-488.
🕮 Facklam, R., & Elliott, J. A. (1995). Identification, classification, and clinical relevance of catalase-negative, gram-positive cocci, excluding the streptococci and enterococci. Clinical microbiology reviews, 8(4), 479-495.
🕮 Gu Liu, C., Green, S. I., Min, L., Clark, J. R., Salazar, K. C., Terwilliger, A. L., & Maresso, A. W. (2020). Phage-antibiotic synergy is driven by a unique combination of antibacterial mechanism of action and stoichiometry. MBio, 11(4), 10-1128.
🕮 Guessennd, N. K., Ouattara, M. B., Nevry, R. K., Gbanon, V., Tiekoura, K. B., & ... & Ger, B. M. (2013). Étude des bactéries multirésistantes des effluents hospitaliers d’un centre hospitalier et universitaire (CHU) de la ville d’Abidjan (Côte d’Ivoire). Journal of Applied Biosciences,, 69, 5456-5464.
🕮 Islam, M. J., Uddin, M. S., Hakim, M. A., Das, K. K., & Hasan, M. N. (2008). Role of untreated liquid hospital waste to the development of antibiotic resistant bacteria. J Innov Dev Strategy, 4(4), 17-21.
🕮 Jarlier, V., Nicolas, M., Fournier, G., & Philippon, A. (1988). Extended-broad spectrum β-lactamases conferring transferable resistance to newer β-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev Infect Dis, 10(4), 867-78.
🕮 Kooli, I., Kadri, Y., Abdallah, H. B., Mhalla, S., Haddad, O., Noomen, S., & Mastouri, M. (2014). Épidémiologie des bactéries multi-résistantes dans une unité néonatale tunisienne. Journal de pédiatrie et de puériculture, 27(5), 236-242.
🕮 Li, S., Ondon, B. S., Ho, S. H., Jiang, J., & Li, F. (2022). Antibiotic resistant bacteria and genes in wastewater treatment plants: From occurrence to treatment strategies. Science of The Total Environment, 156-544.
🕮 Nadella, R. K., Panda, S. K., Badireddy, M. R., Kurcheti, P. P., Raman, R. P., & Mothadaka, M. P. (2022). Multi-drug resistance, integron and transposon-mediated gene transfer in heterotrophic bacteria from Penaeus vannamei and its culture environment. Environmental Science and Pollution Research, 29(25), 37527-37542.
🕮 Parveau, P. (2011). Bacteries multiresistantes dans l’environnement :recherche dans les effluents de la ville de toulouse. Toulouse.
🕮 Reiner, K. (2010). Catalase test protocol. American Society for Microbiology.
🕮 Rennie, R. P., Turnbull, L., Brosnikoff, C., & Cloke, J. (2012). First comprehensive evaluation of the MIC evaluator device compared to Etest and CLSI broth microdilution for MIC testing of aerobic Gram-positive and Gram-negative bacterial species. Journal of clinical microbiology, 50(4), 1147-1152.
🕮 Rodríguez, C. N., Campos, R., Pastran, B., Jimenez, I., Garcia, A., Meijomil, P., & Rodríguez-Morales, A. J. (2005). Sepsis due to extended-spectrum β-lactamase–producing aeromonas hydrophila in a pediatric patient with diarrhea and pneumonia. Clinical infectious diseases, 41(3), 421-422.
🕮 Siu, L., Lu, P., Chen, J., Lin, F., & Chang, S. (2003). High-level expression of AmpC β-lactamase due to insertion of nucleotides between 10 and 35 promoter sequences in Escherichia coli clinical isolates: cases not responsive to extended-spectrumcephalosporin treatment. Antimicrob Agents Chemother, 47(7), 2138-44.
🕮 Taylor, W. I., & Achanzar, D. (1972). Catalase test as an aid to the identification of Enterobacteriaceae. Applied microbiology, 24(1), 58-61.
🕮 Van Twest, R., & Kropinski, A. M. (2009). Bacteriophage enrichment from water and soil. Bacteriophages. Methods and Protocols, Volume 1: Isolation, Characterization, and Interactions, 15-21.
🕮 Varela, A. R., Ferro, G., Vredenburg, J., Yanık, M., Vieira, L., Rizzo, L., & Manaia, C. M. (2013). Vancomycin resistant enterococci: from the hospital effluent to the urban wastewater treatment plant. Science of the Total Environment(450), 155-161.
🕮 Vira, H., Bhat, V., & Chavan, P. (2016). Diagnostic molecular microbiology and its applications: Current and future perspectives. Clin Microbiol Infect Dis, 1(1), 20-31.
🕮 Wenzler, E., Maximos, M., Asempa, T. E., Biehle, L., Schuetz, A. N., & Hirsch, E. B. (2023). Antimicrobial susceptibility testing: An updated primer for clinicians in the era of antimicrobial resistance: Insights from the Society of Infectious Diseases Pharmacists. Pharmacotherapy: The Journal of Human Pharmacology and Drug Therapy, 43(4), 264-278.
Detection of multidrug-resistant bacteria in the effluents of the specialized HAMDANE Bakhta hospital in Saida
HOCINE Kamilia, MEKKATI Khadidja, BENABBOU Taha Ahmed📧
Laboratory of Biotoxicology, Pharmacognosy and biological valorisation of plants, Faculty of Natural and Life Sciences, Department of Biology, University of Saida - Dr Moulay Tahar, 20100 Saida, Algeria.
📧 E. mail: taha.benabbou@univ-saida.dz
Published: 15 January 2024
Abstract
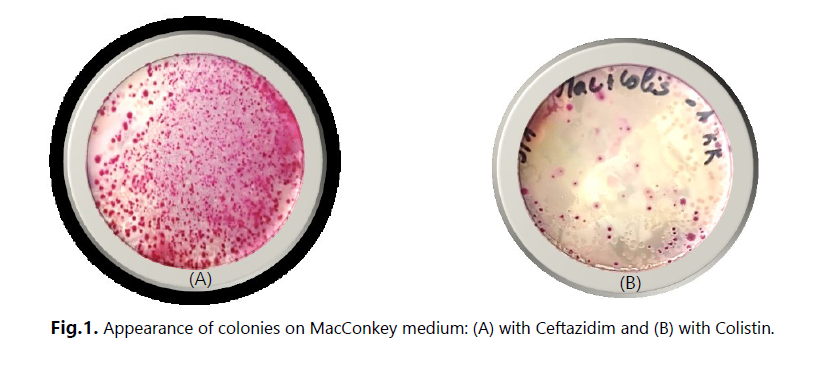
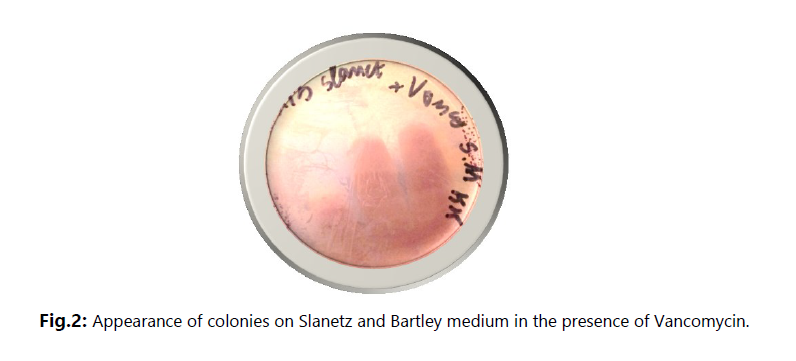
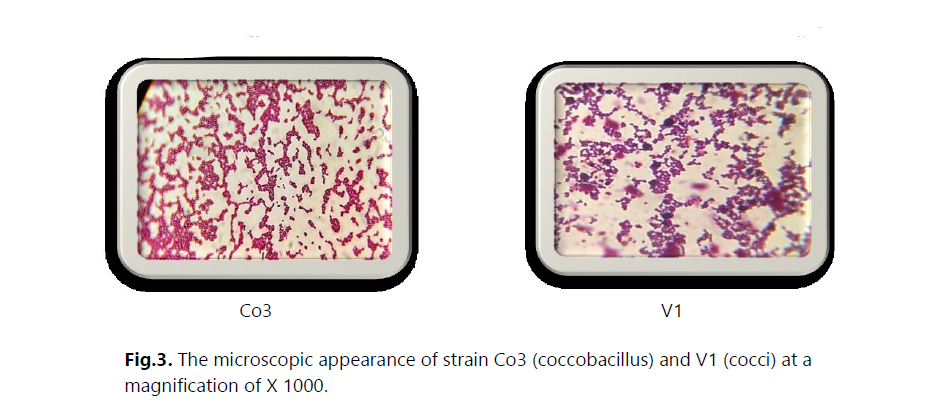
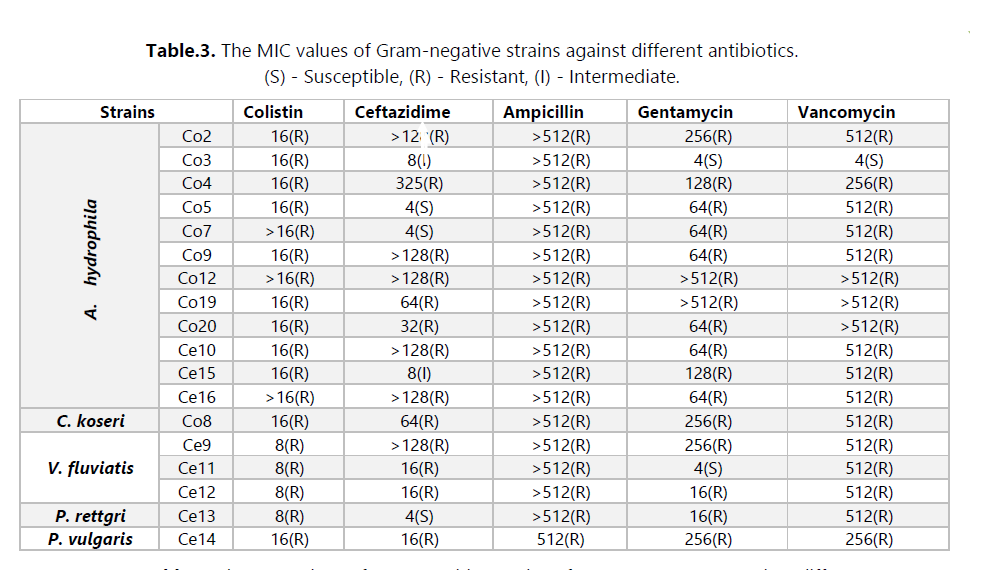
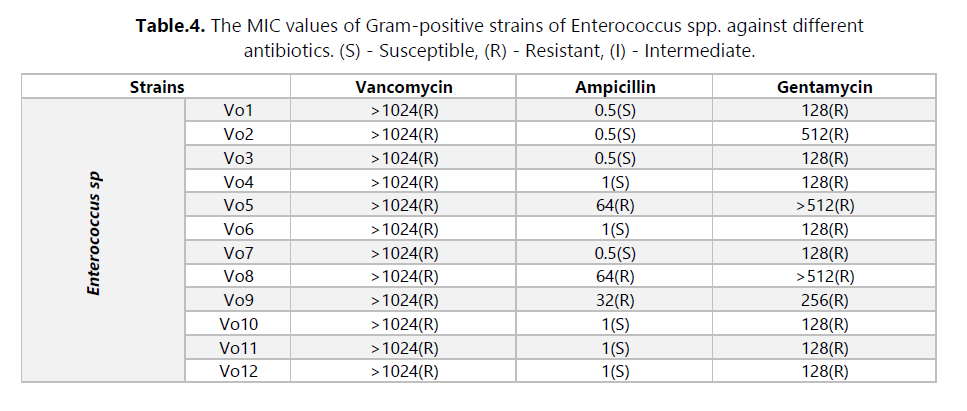
The main objective of this study was to investigate the presence of multidrug-resistant bacteria in untreated hospital effluents from HAMDANE Bakhta Hospital in Saida and evaluate their resistance levels to certain antibiotics. From these hospital effluents, we conducted a count of the resistant flora on MacConkey medium using ceftazidime, revealing a concentration of approximately 2.6 × 106 CFU/mL. Similarly, the colistin-resistant flora was counted at a concentration of 1.4 × 105 CFU/mL, compared to an estimated total flora of around 1 × 107 CFU/mL. Furthermore, the vancomycin-resistant flora on Slanetz and Bertelay medium was present at a concentration of 2.4 × 102 CFU/mL, compared to a total flora concentration of approximately 104 CFU/mL. During our study, we successfully isolated 30 bacterial strains, including 12 strains of Aeromonas hydrophila, 3 strains of Vibrio vulgaris, 1 strain of Citrobacter koseri, 1 strain of Proteus fluviatis, 1 strain of Providencia rettgri, and 12 strains of Enterococcus spp.
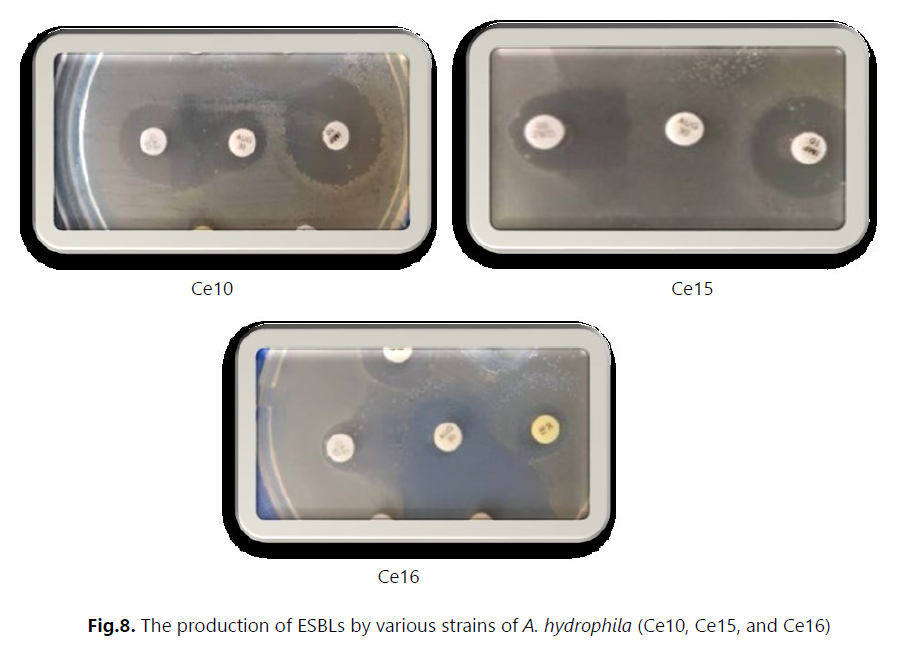
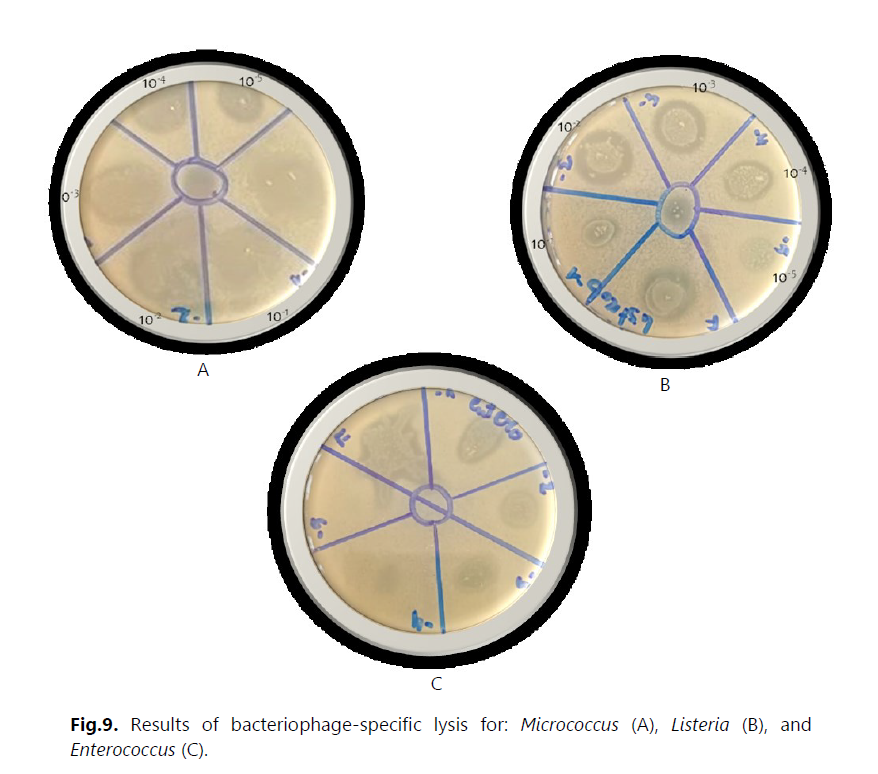
Antibiotic susceptibility was evaluated using the antibiogram and MIC measurement. The results revealed that most strains were multidrug-resistant to various tested antibiotic families, particularly to hospital-used antibiotics such as ceftazidime, colistin, and vancomycin. These strains exhibited a high level of resistance, with MIC values exceeding 512 μg/mL. Additionally, we detected the production of extended-spectrum beta-lactamases (ESBLs) in three strains of Aeromonas hydrophila (Ce10, Ce15, and Ce16). Furthermore, we isolated bacteriophages effective against several bacterial strains, including Staphylococcus, E. coli, Micrococcus, Salmonella, Enterococcus, and others. In this study, we utilized these bacteriophages in combination with ceftazidime. However, no synergy between the two was observed against the Enterococcus strains isolated from hospital effluents.
Multidrug-resistant bacteria have been found in hospital effluents, posing a significant challenge for treating nosocomial infections. Further research is required to develop effective strategies, including exploring alternative combinations of antibiotics, bacteriophages, and other innovative approaches.
Key words
Hospital effluents, antibiotic resistance, ceftazidime, colistin, vancomycin, ESBL, bacteriophages.
I. Introduction
Multiresistant bacteria (MRB) constitute a growing public health problem worldwide, and their presence in hospital effluents is a major concern. These bacteria, often referred to as "superbugs," have developed the ability to resist multiple classes of antibiotics, significantly limiting available treatment options and increasing the risk of complications and death in infected patients (Kooli, et al., 2014).
Hospital effluents, such as wastewater and medical waste, are potential sources of MRB dissemination in the environment. These effluents contain a diversity of microorganisms, including pathogenic bacteria, which may be antibiotic-resistant due to continuous exposure to antimicrobial agents in healthcare facilities (Al Aukidy, Al Chalabi, & Verlicchi, 2018).
MRB isolated from hospital effluents pose a threat to public health in several ways. Firstly, they can be directly transmitted to patients upon admission to the hospital, leading to severe and challenging-to-treat nosocomial infections (Li, Ondon, Ho, Jiang, & Li, 2022). Secondly, these bacteria can be dispersed in the environment, including surface waters, soils, and aquatic ecosystems, contributing to the spread of antibiotic resistance (Guessennd, et al., 2013).
Antibiotic resistance is often associated with the acquisition of resistance genes through horizontal transfer, enabling bacteria to rapidly and efficiently share these resistance mechanisms. Hospital effluents provide a conducive environment for this genetic transfer due to the coexistence of different bacterial species and the presence of mobile genetic elements such as plasmids and integrons (Nadella, et al., 2022).
In this study, our main objective is to characterize multiresistant bacteria isolated from the effluents of HAMDANE Bakhta Hospital. Specifically, we focus on identifying bacteria resistant to last-resort and hospital-use antibiotics, such as ceftazidime, colistin, and vancomycin, in the Saida Wilaya.
To achieve this goal, we are evaluating the antibiotic resistance profile of these bacteria, identifying the antibiotics to which they exhibit resistance. We are particularly interested in antibiotics used in the hospital setting, as resistance to these drugs can have a significant impact on the management of nosocomial infections.
Furthermore, we are exploring solutions to combat the isolated bacteria using a combination approach of phages and antibiotics. Hence, we are investigating the potential of using phages in conjunction with antibiotics to overcome resistance and improve treatment efficacy.
II. Materials and Methods
III. Results and Discussion
IV. Conclusion
Hospital wastewater represents a major source of contamination with antibiotics and multi-drug resistant bacteria, raising significant concerns regarding public health. These wastewater streams from hospitals, rich in organic matter, create an aquatic environment conducive to bacterial proliferation. Among these microorganisms, Gram-negative bacilli such as Enterobacteriaceae, Vibrionaceae, and Aeromonadaceae, as well as Gram-positive cocci like Enterococcaceae, play a substantial role in human infectious diseases due to their antibiotic resistance.
In our study, we focused on the search for multi-drug resistant bacteria in the effluents of HAMDANE Bakhta Hospital, located in the Saida province. Among the different bacterial species identified, we observed a high frequency of A. hydrophila presence in these effluents, suggesting a probable origin of these microorganisms within the hospital environment. We also detected other species such as P. rettgeri, P. vulgaris, C. koseri, V. fluvialis, and Enterococcus spp.
The results obtained revealed high resistance to several antibiotics, particularly amoxicillin plus clavulanic acid (AUG) and hospital-use antibiotics such as ceftazidime, colistin, and vancomycin, with extremely high minimum inhibitory concentrations (MICs). On the other hand, imipenem appears to exhibit some effectiveness against these bacterial strains. For example, resistance to beta-lactams may be attributed to the production of enzymes such as extended-spectrum beta-lactamases (ESBLs) produced by A. hydrophila strains.
The susceptibility to other antibiotics varies depending on each bacterial family. It is important to note that these results are based on available data, and a more in-depth analysis would require a better understanding of bacterial strains, their resistance mechanisms, and the history of antibiotic use in the hospital environment.
In the face of this concerning situation, it is essential to implement effective preventive and control measures. One promising approach that we have explored is the combined use of phages and antibiotics to combat multi-drug resistant bacteria. Phages are bacteria-specific viruses capable of selectively targeting and eliminating these microorganisms. By combining phages with antibiotics, we could enhance treatment efficacy and reduce the spread of resistant bacteria.
The preliminary results obtained in this study open up several avenues for further research. It would be relevant to expand our study to a larger geographical area over an extended period, perform strain identification using molecular and serological techniques, characterize the presence of extended-spectrum beta-lactamases (ESBLs) using PCR, and study the nationwide impact of these effluents on the environment.
References
🕮 Al Aukidy, M., Al Chalabi, S., & Verlicchi, P. (2018). Hospital wastewater treatments adopted in Asia, Africa, and Australia. Hospital wastewaters: characteristics, management, treatment and environmental risks, 171-188.
🕮 Baumont, S. (2004). Réutilisation des eaux usées épurées : risques et faisabilité en Ile de France. Toulouse.
🕮 Bradford, P. A. (2001). Extended-spectrum β-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clinical microbiology reviews, 14(1), 933-951.
🕮 CLSI. (2012). Performance Standards for Antimicrobial Susceptibility Testing. Clinical and Laboratory Standards Institute. 26th Edition, M100S, Wayne. USA.
🕮 Cockerill, F. R. (2012). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow. Approved Standard—Ninth Edition. CLSI, 12.
🕮 Ekhaise, F., & Omavwoya, B. P. (2008). Influence of hospital wastewater discharged from University of Benin Teaching Hospital (UBTH), Benin City on its receiving environment. American-Eurasian J Agric Environ Sci,, 4(4), 484-488.
🕮 Facklam, R., & Elliott, J. A. (1995). Identification, classification, and clinical relevance of catalase-negative, gram-positive cocci, excluding the streptococci and enterococci. Clinical microbiology reviews, 8(4), 479-495.
🕮 Gu Liu, C., Green, S. I., Min, L., Clark, J. R., Salazar, K. C., Terwilliger, A. L., & Maresso, A. W. (2020). Phage-antibiotic synergy is driven by a unique combination of antibacterial mechanism of action and stoichiometry. MBio, 11(4), 10-1128.
🕮 Guessennd, N. K., Ouattara, M. B., Nevry, R. K., Gbanon, V., Tiekoura, K. B., & ... & Ger, B. M. (2013). Étude des bactéries multirésistantes des effluents hospitaliers d’un centre hospitalier et universitaire (CHU) de la ville d’Abidjan (Côte d’Ivoire). Journal of Applied Biosciences,, 69, 5456-5464.
🕮 Islam, M. J., Uddin, M. S., Hakim, M. A., Das, K. K., & Hasan, M. N. (2008). Role of untreated liquid hospital waste to the development of antibiotic resistant bacteria. J Innov Dev Strategy, 4(4), 17-21.
🕮 Jarlier, V., Nicolas, M., Fournier, G., & Philippon, A. (1988). Extended-broad spectrum β-lactamases conferring transferable resistance to newer β-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev Infect Dis, 10(4), 867-78.
🕮 Kooli, I., Kadri, Y., Abdallah, H. B., Mhalla, S., Haddad, O., Noomen, S., & Mastouri, M. (2014). Épidémiologie des bactéries multi-résistantes dans une unité néonatale tunisienne. Journal de pédiatrie et de puériculture, 27(5), 236-242.
🕮 Li, S., Ondon, B. S., Ho, S. H., Jiang, J., & Li, F. (2022). Antibiotic resistant bacteria and genes in wastewater treatment plants: From occurrence to treatment strategies. Science of The Total Environment, 156-544.
🕮 Nadella, R. K., Panda, S. K., Badireddy, M. R., Kurcheti, P. P., Raman, R. P., & Mothadaka, M. P. (2022). Multi-drug resistance, integron and transposon-mediated gene transfer in heterotrophic bacteria from Penaeus vannamei and its culture environment. Environmental Science and Pollution Research, 29(25), 37527-37542.
🕮 Parveau, P. (2011). Bacteries multiresistantes dans l’environnement :recherche dans les effluents de la ville de toulouse. Toulouse.
🕮 Reiner, K. (2010). Catalase test protocol. American Society for Microbiology.
🕮 Rennie, R. P., Turnbull, L., Brosnikoff, C., & Cloke, J. (2012). First comprehensive evaluation of the MIC evaluator device compared to Etest and CLSI broth microdilution for MIC testing of aerobic Gram-positive and Gram-negative bacterial species. Journal of clinical microbiology, 50(4), 1147-1152.
🕮 Rodríguez, C. N., Campos, R., Pastran, B., Jimenez, I., Garcia, A., Meijomil, P., & Rodríguez-Morales, A. J. (2005). Sepsis due to extended-spectrum β-lactamase–producing aeromonas hydrophila in a pediatric patient with diarrhea and pneumonia. Clinical infectious diseases, 41(3), 421-422.
🕮 Siu, L., Lu, P., Chen, J., Lin, F., & Chang, S. (2003). High-level expression of AmpC β-lactamase due to insertion of nucleotides between 10 and 35 promoter sequences in Escherichia coli clinical isolates: cases not responsive to extended-spectrumcephalosporin treatment. Antimicrob Agents Chemother, 47(7), 2138-44.
🕮 Taylor, W. I., & Achanzar, D. (1972). Catalase test as an aid to the identification of Enterobacteriaceae. Applied microbiology, 24(1), 58-61.
🕮 Van Twest, R., & Kropinski, A. M. (2009). Bacteriophage enrichment from water and soil. Bacteriophages. Methods and Protocols, Volume 1: Isolation, Characterization, and Interactions, 15-21.
🕮 Varela, A. R., Ferro, G., Vredenburg, J., Yanık, M., Vieira, L., Rizzo, L., & Manaia, C. M. (2013). Vancomycin resistant enterococci: from the hospital effluent to the urban wastewater treatment plant. Science of the Total Environment(450), 155-161.
🕮 Vira, H., Bhat, V., & Chavan, P. (2016). Diagnostic molecular microbiology and its applications: Current and future perspectives. Clin Microbiol Infect Dis, 1(1), 20-31.
🕮 Wenzler, E., Maximos, M., Asempa, T. E., Biehle, L., Schuetz, A. N., & Hirsch, E. B. (2023). Antimicrobial susceptibility testing: An updated primer for clinicians in the era of antimicrobial resistance: Insights from the Society of Infectious Diseases Pharmacists. Pharmacotherapy: The Journal of Human Pharmacology and Drug Therapy, 43(4), 264-278.