JSBB: Volume 2, Issue 1, June 2023 - ANTIBIOTICS RESISTANCE
Concept article.
Antibiotic Resistance and the Ribosome:
A key Target in the Battle against Pathogens
A key Target in the Battle against Pathogens
RACHEDI Abdelkrim
Laboratory of Biotoxicology, Pharmacognosy and biological valorisation of plants, Faculty of Sciences, Department of Biology, University of Saida - Dr Moulay Tahar, 20100 Saida, Algeria.
📧 E. mail: abdelkrim.rachedi@univ-saida.dz
Published: 09 June 2023
Abstract
Antibiotic resistance poses a significant global threat to public health, necessitating innovative
strategies to combat multidrug-resistant pathogens. In here we focuses on the ribosome as a key
target in the fight against antibiotic resistance.
Ribosomes, can be referred to as the "ultimate molecular 3D printers", of the flesh and blood, are ancient molecular machines
responsible for protein synthesis. Pathogenic bacteria heavily rely on ribosomes for survival and proliferation.
By disrupting ribosomal function, researchers aim to hinder bacterial protein synthesis, effectively impeding
bacterial growth and reducing the spread of antibiotic-resistant strains. Targeting the ribosome offers a
promising avenue for the development of novel antibiotics that can overcome resistance mechanisms employed
by bacteria.
Ongoing research endeavors seek to understand the intricacies of ribosomal structure and function, identifying
vulnerabilities that can be exploited for the design of more effective and specific drugs. We explore, in this article,
the significance of ribosomes in biological processes and highlights the ongoing efforts to leverage ribosomal
targeting in combating superbugs. Scientists aim to mitigate antibiotic resistance and safeguard public health through
gaining insights into ribosomal mechanisms and advancing intervention strategies.
Key words
Antibiotic resistance, Ribosome, Protein synthesis, Multidrug-resistant pathogens, Superbugs, Drug development.
Introduction
Antibiotic resistance is an increasingly urgent global concern, undermining the effectiveness of existing
antimicrobial therapies and posing a substantial threat to public health (Wilson, 2014). The emergence of
multidrug-resistant pathogens has necessitated the development of innovative approaches to combat this
critical problem. One promising avenue of research lies in targeting the ribosome, an ancient and essential
molecular machine responsible for protein synthesis. Ribosomes are present in all
living organisms and are often described as the "ultimate molecular 3D printers", of the flesh and blood, due
to their ability to convert genetic information into specific three-dimensionally structured proteins that
execute vital specialized biological functions.
Pathogenic bacteria, which cause a range of infectious diseases, rely heavily on ribosomes for their survival
and proliferation. Disrupting the protein synthesis machinery within bacterial ribosomes
can have a profound impact on bacterial growth, making ribosomes an attractive target for the development of
new antibiotics (Jinzhong et al., 2018). Through interfering with ribosomal function, researchers aim to inhibit
bacterial protein synthesis, thereby halting bacterial growth and reducing the spread of antibiotic-resistant strains.
Targeting the ribosome represents a promising avenue for combating antibiotic resistance due to its fundamental
role in biological processes. Ribosomes are conserved across diverse bacterial species,
making them an ideal target for the design of antibiotics that can circumvent resistance mechanisms employed by
bacteria (Wilson, 2014). Scientists and researchers are actively studying the structure and function of ribosomes,
Figures 1 and 3, to gain a comprehensive understanding of their mechanisms. Unravelling the intricacies of ribosomal biology can
lead to the identification of vulnerabilities that can be exploited in the development of drugs with greater specificity
and efficacy.
Of particular concern are superbugs, highly resistant bacterial strains that pose significant challenges in
healthcare settings. Ribosomes have been found to play a key role in combating superbugs, and ongoing
research efforts are focused on harnessing the potential of ribosomal targeting to overcome antibiotic
resistance (Science Museum blog). Researchers aim at gaining deeper insights into ribosomal mechanisms and
advancing intervention strategies, to mitigate the spread of multidrug-resistant pathogens and protect public health.
This article delves into the significance of ribosomes in biological processes, highlighting their role as a
focal point in the fight against antibiotic resistance. It explores ongoing research and advancements in
ribosomal targeting, aiming to shed light on the potential of ribosome-based interventions in combating
superbugs and overcoming antibiotic resistance. Scientists strive to develop effective strategies to safeguard public
health and preserve the efficacy of antibiotic therapies by focusing on this critical area of study.
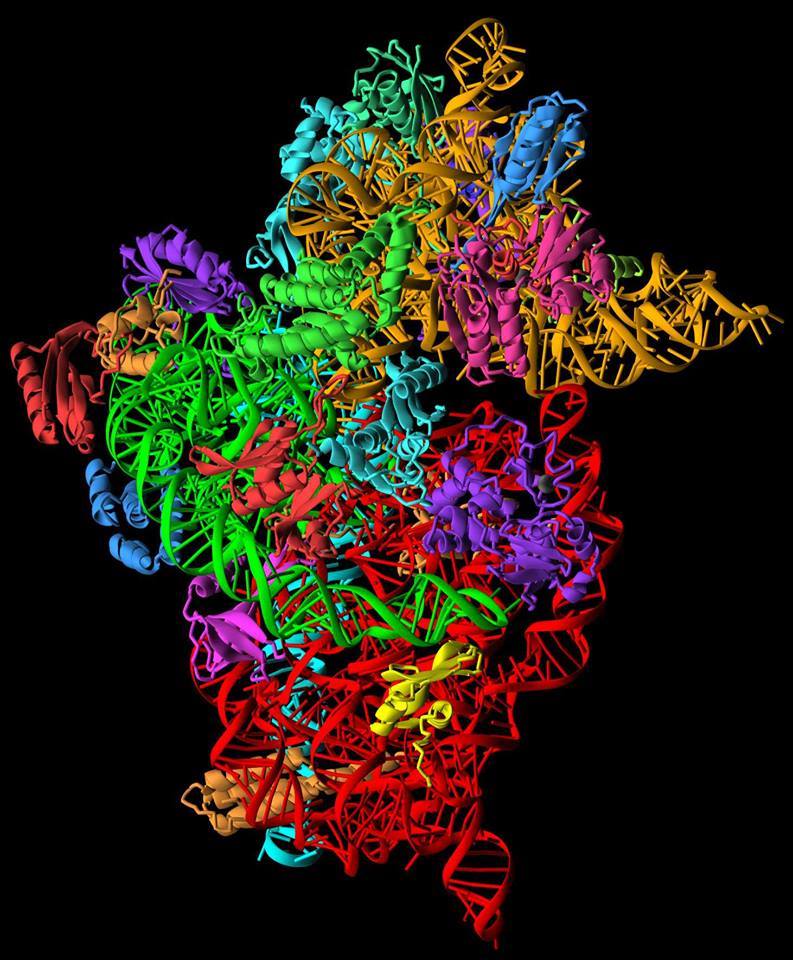
Figure 1. The 30S component, or ‘brain’ of the ribosome.
Credit: Ramakrishnan Lab. https://blog.sciencemuseum.org.uk/ultimate-molecular-machine-plays-key-role-in-superbug-fight
Credit: Ramakrishnan Lab. https://blog.sciencemuseum.org.uk/ultimate-molecular-machine-plays-key-role-in-superbug-fight
The Ribosome; an Ultimate Molecular 3D Printer
The ribosome, often referred to as the "ultimate molecular 3D printer of flesh and blood" is a complex cellular structure
found in all living organisms. It acts as a translation machine, converting genetic information encoded
in RNA molecules into functional proteins with specific structures necessary for the correct biology to happen.
Ribosomes accomplish this through linking together amino acids in the precise order dictated by the genetic code, Figure 2.
Figure 2. Animated model of protein synthesis by a Ribosome found in bacteria
Credit: Ramakrishnan Lab. https://blog.sciencemuseum.org.uk/wp-content/uploads/2016/05/ezgif.com-video-to-gif.gif
Credit: Ramakrishnan Lab. https://blog.sciencemuseum.org.uk/wp-content/uploads/2016/05/ezgif.com-video-to-gif.gif
Ribosomes and Pathogenic Bacteria
Ribosomes play a critical role in the life cycle of pathogenic bacteria, which are responsible for causing various
infectious diseases. These bacteria heavily rely on ribosomes for their survival and proliferation, making them an
attractive target for therapeutic interventions. Disrupting the ribosomal machinery can impede
the ability of pathogenic bacteria to synthesize proteins, leading to a significant impact on their growth and
proliferation (Jinzhong et al., 2018).
One approach to targeting ribosomes is the development of ribosome-targeted antibiotics.
These antibiotics specifically inhibit the activity of bacterial ribosomes, selectively
disrupting protein synthesis in pathogenic bacteria while sparing the ribosomes of host cells.
These antibiotics would selectively target the bacterial ribosomes and can effectively inhibit
bacterial growth and prevent the spread of infections caused by resistant bacteria.
The advantage of targeting ribosomes lies in their conserved structure and function across different
bacterial species. Ribosomes are ancient molecular machines that share fundamental characteristics,
allowing researchers to develop broad-spectrum antibiotics that can combat a wide range of pathogenic
bacteria. This broad-spectrum activity is particularly valuable in the face of
antibiotic resistance, as it provides a versatile approach to combating resistant strains.
In recent years, research efforts have focused on gaining a deeper understanding of ribosomal structure,
function, and the mechanisms by which ribosome-targeted antibiotics work. The identification of specific
vulnerabilities that can be exploited for the design of more effective drugs is potentially within reach through
understanding the intricate details of ribosomal biology. This knowledge has paved the way for the development
of novel ribosome-targeted antibiotics with enhanced potency, reduced side effects, and improved resistance
profiles.
The Fight against Antibiotic Resistance
As bacteria evolve and develop resistance mechanisms against conventional antibiotics, finding alternative
strategies to combat them becomes crucial. Targeting the ribosome presents a promising avenue, as it
represents a fundamental and conserved biological process across diverse bacterial species.
Research projects working on interfering with ribosomal function would potentially result in inhibiting bacterial protein synthesis
and ultimately halting bacterial growth and reduce the spread of antibiotic-resistant strains (Wilson, 2014).
Ribosome-targeted antibiotics work by binding to specific sites within the ribosome, inhibiting its function
and disrupting the accurate synthesis of proteins. This interference with protein synthesis compromises the
survival and replication of bacteria, effectively reducing their ability to cause infections (Wilson, 2014).
Moreover, targeting the ribosome has the potential to overcome certain resistance mechanisms employed by bacteria.
While bacteria can develop resistance to antibiotics through various mechanisms, such as modifying drug targets
or enhancing efflux pumps, the ribosome represents a fundamental process that is difficult for bacteria to evade
without severely impairing their own survival. Targeting the ribosome allowed researchers
to circumvent some of the common resistance mechanisms employed by bacteria, making ribosome-targeted antibiotics
a promising strategy in combating antibiotic-resistant strains.
Research and Advancements
Advancements in technology and techniques, such as cryo-electron microscopy and X-ray crystallography,
have revolutionized the field of ribosomal research. These techniques enable researchers to visualize
the three-dimensional structure of ribosomes at high resolution, providing insights into their molecular
architecture and functional mechanisms, see the Figures 3 & 4. Scientific research in analysing these
structures, would pinpoint specific sites within the ribosome that are critical for protein synthesis and
can potentially be targeted by antibiotics.
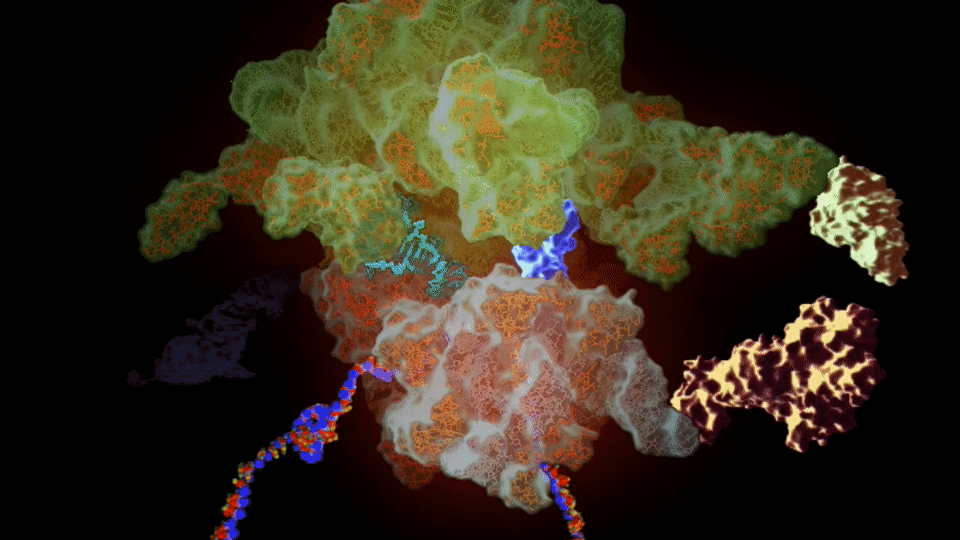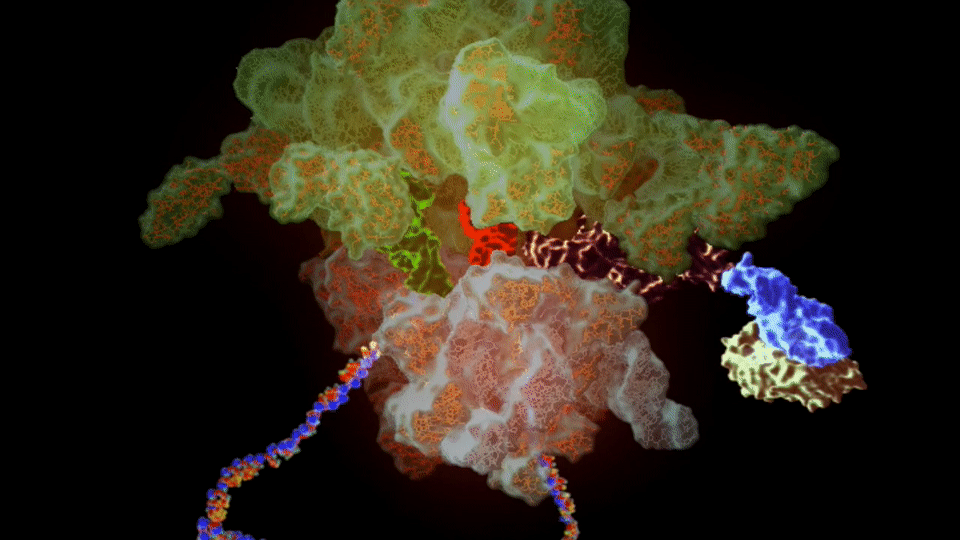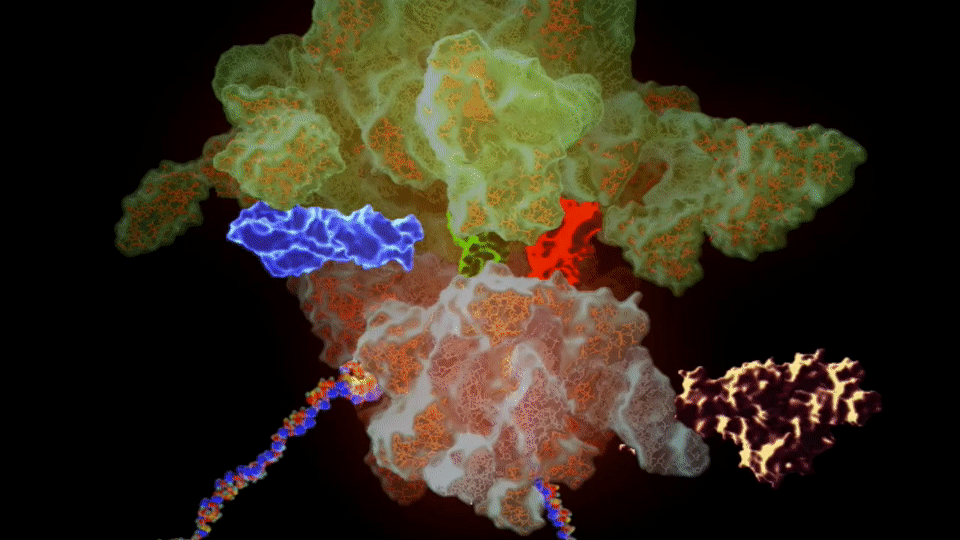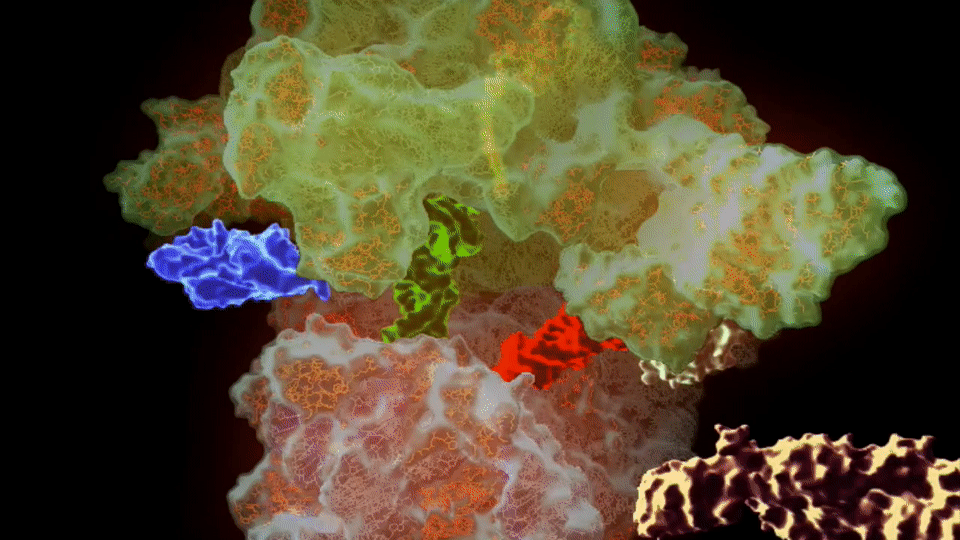
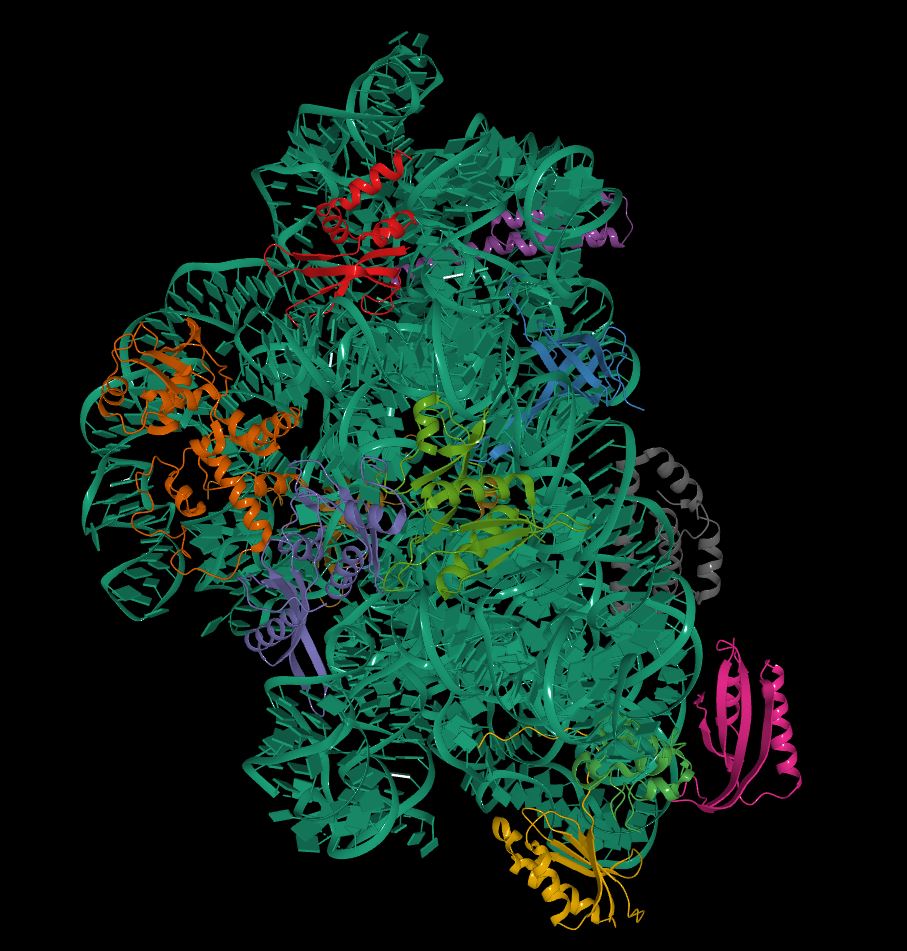
Figure 3. Cryo-Electron Microscopy Structure of The 70s ribosome of Enterococcus Faecalis.
Source: the Protein Databank (PDB) entry code 6WUB; can be explored using the SSFS tool
https://bioinformaticstools.org/ssfs/ssfs.php?qry=6WUB,
see next figure.
Source: the Protein Databank (PDB) entry code 6WUB; can be explored using the SSFS tool
https://bioinformaticstools.org/ssfs/ssfs.php?qry=6WUB,
see next figure.
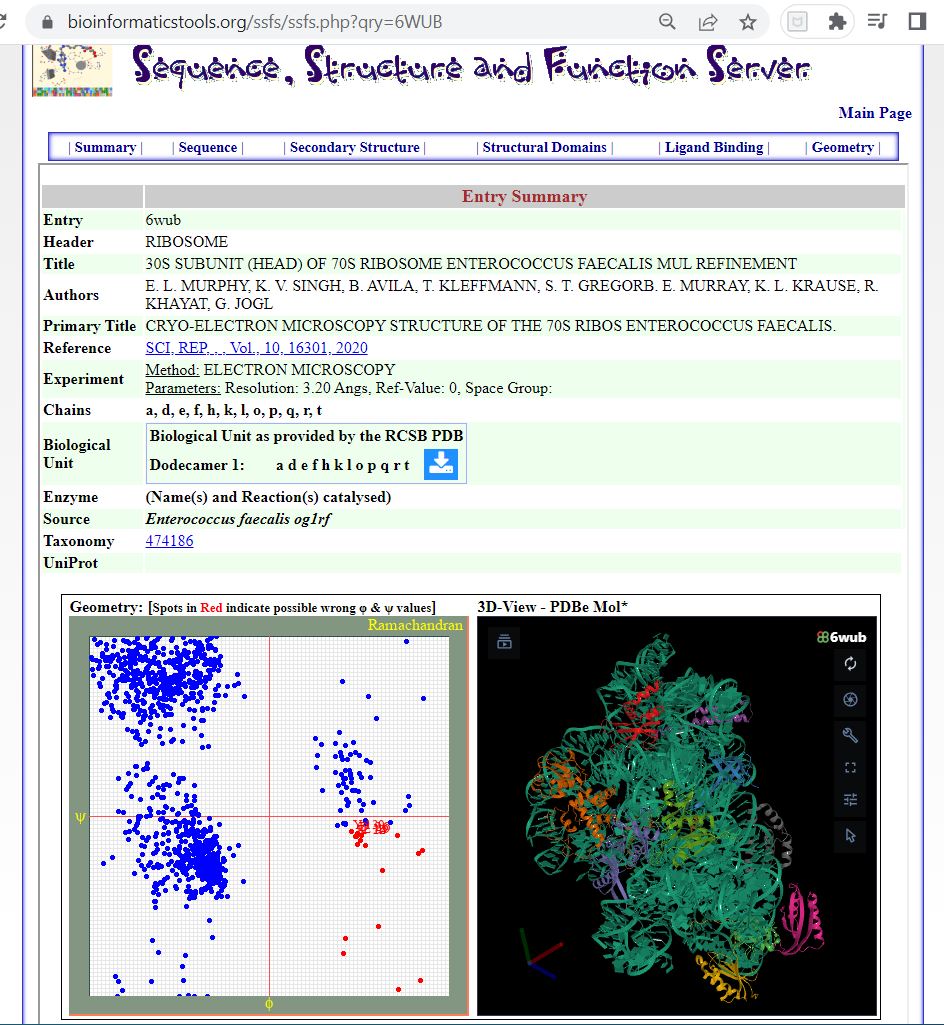
Figure 4. The SSFS❇️ tool showing important details of the PDB entry 6WUB related to the 30S Ribosome structure.
https://bioinformaticstools.org/ssfs/ssfs.php?qry=6WUB.
https://bioinformaticstools.org/ssfs/ssfs.php?qry=6WUB.
❇️ SSFS: Sequence, Structure and Function Server by the Structural Biology & Bioinformatics Groups, Biology Dept, Faculty of Science, University of Saida, Algeria
نظام: التركيب الأولي، التركيب الفراغي و الوظيفة البيولوجية ، فريق البيولوجيا ثلاثية
الأبعاد و المعلوماتية الحيوية، قسم البيولوجيا، كلية العلوم، جامعة سعيدة، الجزائر
Moreover, studying the ribosome allows researchers to understand the mechanisms by which ribosome-targeted
antibiotics interact with the ribosome and inhibit protein synthesis. This knowledge aids in the design and
optimization of antibiotics that can effectively bind to the ribosome and disrupt its function, thereby
halting bacterial growth.
One area of focus in ribosomal research is the development of antibiotics that exhibit high specificity for
bacterial ribosomes while sparing eukaryotic ribosomes found in human cells. This selectivity is crucial to
minimize potential side effects and toxicity to the host organism. Understanding the subtle differences
between bacterial and human ribosomes, can help in the design endeavours of novel antibiotics that selectively
target bacterial ribosomes, enhancing their efficacy and safety profile.
A number of research projects are investigating novel mechanisms of action for ribosome-targeted antibiotics.
Exploration of the different modes of interference with ribosomal function, such as inhibition of specific
steps in the protein synthesis process or disruption of ribosome assembly, scientists aim also to expand the
repertoire of antibiotics and overcome existing resistance mechanisms.
Furthermore, advancements in computational modelling and virtual screening techniques have facilitated
the identification and optimization of potential ribosome-targeted antibiotics. Computational biology and
computer simulations in addition to large-scale virtual screening of compound libraries, would result in vast
number of molecules and predict their binding affinity and potential efficacy against the ribosome.
This computational approach accelerates the discovery and development of novel antibiotics, potentially
shortening the timeline for bringing new drugs to the market.
The link between Ribosomes and Superbugs
Superbugs, referring to highly resistant bacterial strains, pose a significant challenge in healthcare settings.
Ribosomes have been found to play a key role in combating superbugs, and ongoing research efforts are focused on
harnessing the potential of ribosomal targeting to overcome antibiotic resistance (Science Museum blog).
For further information on this topic, the article "The Ultimate Molecular Machine Plays Key Role in Superbug Fight"
(source: Science Museum blog) offers a comprehensive overview.
Conclusion
The fight against antibiotic resistance requires innovative approaches that target the fundamental
processes essential for bacterial survival. The ribosome, as the ancient molecular machine responsible
for protein synthesis, has emerged as a crucial focal point in this battle.
Understanding the intricate workings of ribosomes and developing targeted interventions, researchers
aim to overcome antibiotic resistance and safeguard public health. Continued research and advancements
in this field hold great promise for the development of effective strategies to combat
multidrug-resistant pathogens.
References
🕮 Jinzhong L, Dejian Z, Thomas AS, Yury SP, Matthieu GG. (2018). Ribosome-targeting antibiotics: mode of action, mechanisms of resistance, and implications for drug design. Annu Rev Biochem. Jun 20;87:451-478. doi: 10.1146/annurev-biochem-062917-011942. Available at: https://pubmed.ncbi.nlm.nih.gov/29570352/
🕮 Wilson DN. (2014). Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat Rev Microbiol. 12(1):35-48. doi: 10.1038/nrmicro3155. Available at: https://www.nature.com/articles/nrmicro3155
🕮 Wimberly BT, Brodersen DE, Clemons WM Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. (2000). Structure of the 30S ribosomal subunit. Nature.407(6802):327-39. doi: 10.1038/35030006. PMID: 11014182. Available at: https://www.nature.com/articles/35030006
🕮 Mishra S, Imlay JA. (2012) Why do bacteria use so many enzymes to scavenge hydrogen peroxide? Arch Biochem Biophys. 525(2):145-60. doi: 10.1016/j.abb.2012.02.028. PMID: 22405803. Available at: https://pubmed.ncbi.nlm.nih.gov/22609271/
🕮 Benabbou TA, Benreguieg M, Bellil Y, Chahrour W (2023). Antibiotic Resistance: A Global Public Health Crisis and Current Strategies for Combatting It. JSBB 2(1). Available at: https://bioinformatics.univ-saida.dz/jsbb/blog-single.php?ar=April_2023LOLA_Global_Public_Health_Crisis_and_Current_Strategies_for_Combatting_It
🕮 Rachedi A (2023). Teixobactin an antibiotic from soil bacteria for fighting Multiple Antibiotic Resistance phenomena. JSBB 2(1). Available at: https://bioinformatics.univ-saida.dz/jsbb/blog-single.php?ar=April_2023LOLTeixobactin_Soil_Antibiotic_4_Multiple_Antibiotic_Resistance
🕮 Science Museum blog. "The Ultimate Molecular Machine Plays Key Role in Superbug Fight." Available at: https://blog.sciencemuseum.org.uk/ultimate-molecular-machine-plays-key-role-in-superbug-fight/