JSBB: Volume 2, Issue 4, March 2024 - ANTIBIOTICS RESISTANCE ARTICLES,
Research article: Master's research based.
BARID database: Data Annotation and Integration of Experimental Investigation of Bacterial Antibiotic Resistance and Antibacterial Activity of Annona cherimola Mill. Seeds
KHERIS Soumeya1, BENREGUIEG Mokhtar1 and RACHEDI Abdelkrim1,📧
1 Departement of Biology, Faculty of Natural and Life Sciences, University of Saida-Dr. Moulay Tahar, 20000 Saida, Algeria
2 Laboratory of Biotoxicology, Pharmacognosy and biological valorisation of plants Compus Ain El Hadjar, Faculty of Sciences, Department of Biology, University of Saida Dr Tahar Moulay, 20100 Saida, Algeria.
📧 RACHEDI Abdelkrim - E. mail: abdelkrim.rachedi@univ-saida.dz
Published: 15 March 2024
Abstract
The Bacterial Antibiotic Resistance Investigation Database, BARID, is a database that annotates and stores information pertaining to mainly empirical data representing laboratory experiments done on 5 important bacteria known to have developed Antibiotic Resistance; a current medical problem dangerously threatening the health and even human life on global scale.
The bacteria namely Staphylococcus aureus, Clostridium difficile, Escherichia coli, Pseudomonas aeruginosa and Klebsiella pneumoniae which are responsible for a myriad of human diseases where growth tested against 14 commonly used antibiotics belonging to a number of class including Beta-Lactams, Aminoglycosides, Tetracyclines, Macrolides, Chloramphenicol, and Quinolone and the relatively new group of drugs class Oxazolidinones. Moreover, the database annotated the antibacterial activity of Cherimoya fruit seeds extracts on the listed above bacteria. Cherimoya plant has been used in the study as a model of medicinal plant.
In addition to annotating such medically important data, the BARID database offers a number of features based on Datamining and Integration setting the grounds for providing researchers with even deeper insights on the genomic and proteomic causes might be behind the phenomena of antibiotics resistance. More insights could be provided by th 3D-structures, drugs data and scientific literature included in the search outputs of the database.
The BARID database, through a number of novel exploration features, presents users with a set of results including which bacteria are resistant to which antibiotic and which bacteria are sensitive to which of the four types of Cherimoya seeds extracts tested in the study. The system, as it further develops, may potentially also be used to suggest new natural products, such as the extracts of the Cherimoya seeds as alternatives to antibiotics in the cases where antibiotic resistance is shown.
Availability: BARID database is available online at the link: https://www.bioinformaticstools.org/barid/
Keywords: Bacterial Antibiotic Resistance, Anti-Bacterial Activity, Disease, Database, Annotation, Datamining, Data Integration, Alternative natural drugs.
Introduction
Antibiotic resistance is a form of drug resistant whereby some sub-populations of a microorganism, usually a bacterial species, are able to survive after exposure to one or / more antibiotics. In other word, the term “antibiotic resistance” refers to the ability of a microorganism to withstand the effect of an antibiotic. The use of antibiotics is limited because bacteria have evolved defences against certain antibiotics. One of the main mechanisms of defence is inactivation of the antibiotic. This is the usual defence against penicillin and chloramphenicol, among others. Another form of defence involves a mutation that changes the bacterial enzyme affected by the drug in such a way that the antibiotic can no longer inhibit it. This is the main mechanism of resistance to the compounds that inhibit protein synthesis, such as the tetracycline
The problem of resistance has been exacerbated by the use of antibiotics as prophylactics, intended to prevent infection before it occurs. Indiscriminate and inappropriate use of antibiotics for the treatment of the common cold and other common viral infections, against which they have no effect, removes antibiotic-sensitive bacteria and allows the development of antibiotic-resistant bacteria (Witte, 2004)
The pathogens resistant to multiple antibiotics are considered as Multi-drug resistant pathogens (D'Costa, 2011). Some of the most ten important pathogens types of multiple drug resistant organisms that have been encountered include; Staphylococcus aureus, Klebsilla pneumonia, Escherichia coli, Pseudomonas aeruginosa, Nesseria gonhoreae, Clostridium difficile, Mycobacterium tuberculosis, Burkhoderia cepacia, Acenitobacter baumanii, and Streptococcus pyogenes.
In realising the antibiotic resistance as a major problem in the treatment of bacterial infections, there is need to find alternative ways of treating infectious diseases using medicinal plants that are abundantly available and their extracts may overcome the antibiotic resistance that can serve as a source of novel drugs for the treatment of these diseases.
Annona cherimola Mill. known as Cherimoya or Cherimola (Anna Perrone et. al., 2022) is one of the most important fruits of the Annonaceae family known to botanists as one of the three best fruits in the world (National Research Council, 1989; Gardiazbal and Rosenberg, 1993). The famous 18th century Czech naturalist Thaddus Haenke considered it "the masterpiece of Nature". The abbreviation "Mill.", in the scientific name of the Cherimoya plant, stands for the botanist who first described the species, in this case, Philip Miller.
In this age of informatics highly reliance, computers and databases are used to help in solving many problems in biology associated with disease fighting, numerical analysis of complex data generated by the biologists which resulted in the creation of the whole field of science become known as Bioinformatics.
A huge amount of data have been generated in relation to the Antibiotic Resistance and Antibacterial activity experiments carried out in this research. Annotation, data integration and database programming techniques have been implemented to classify, store, analyse the data. Methods have also been devised for query the data and help retrieve potentially relevant research results and display them in meaningful manner to interested users.
Materials and Methods
Bacterial Material:
Five bacterial strains reported antibiotic resistance were available for use in this project: Escherichia coli (ATCC 25922), Staphylococcus aureus (ATCC 25923), Pseudomonas aeruginosa (ATCC 27853), Klebsiella pneumonia (ATCC 25923) according to ’the American type culture collection (ATCC). The bacterial strains were obtained from the Department of cellular and molecular biology, Faculty of Science, University of Telemcen.
Two bacterial strains; Clostridium difficile and Mycobacterium tuberculosis were isolated locally in standard hygienic conditions at the microbiology laboratory of Saida University. However, work and results done on the bacterium Mycobacterium tuberculosis are not included in this version of the BARID database.
The laboratory confirmation of bacterial stains from stock bacterial cultures is reported in master research work successfully approved by jury of scientists (Soumeya Kheris., 2019).
Antibiotics products:
The antibiotics used were chosen to be from the list of the commonly and usually given by local doctors to treat patients with illnesses cases thought caused by the five bacteria mentioned above.
The antibiotics used included the following: Oxaciline, Ampiciline, Amoxiciline, Cefazolin, Ceftazidime, Gentamicine, Amikacin, Kanamycin, Tetracycline, Speramicine, Linezolid, Chloramphenecole, Levofloxin, Nitroxoline. (Soumeya Kheris., 2019).
Plant Material:
The ripen fruits of Annona cherimolla (mill), commonly known as Cherimoya, were imported from spain and obtained from local market in Saida city, Algeria. Cherimoya seeds were harvested from the fresh ripen fruit. The seeds were then separated from the pulp (Figure 1), then dried up under a shade area at room temperature to guarantee total drying and to prevent the breakdown of pharmacological elements (Figure 2).
 Figure1. Seeds still in the pulp flesh of the Cherimoya fruit before harvesting and drying. |
 Figure2. Cherimoya seeds after drying. |
The dried out seeds were then milled into a fine powder using an electric grinder (Figure 3) The powdered material was put in sealed glass bottle and stored in cold room. (Soumeya Kheris., 2019)
Data Preparation & Programing Tools:
A number of software tools were implemented in the realization of the bioinformatics part of this project included the following:
-
Microsoft Excel 2010 (Microsoft Corporation, 2018) has been used in creating all of the histograms and graphs related to the statistical analysis of the web-lab results. It has also been used for data formatting and preliminary storing prior to the database creation.
Results
As reported in the previous chapter, the data generated by the experimental part of this project, outlaid above, have been processed and stored in an in-house database (development database). The in-house database has been further developed into an operational one and made available online. The following text presents the online database, its different way of usage and discussion.
The Online Database:
The production database, named in the project as BARID for Bacterial Antibiotic Resistance Investigation Database has been mounted on a web-server to provide the data and results to local and international community of researcher including students.
The database is available online via the web-address:
https://www.bioinformaticstools.org/barid/
Web access and Querying:
The main interface page of the database, in Figure 4, and highlighted are the main three different ways of querying; Bacteria-tab, Antibiotics-tab and Cherimoya Extracts-tab.
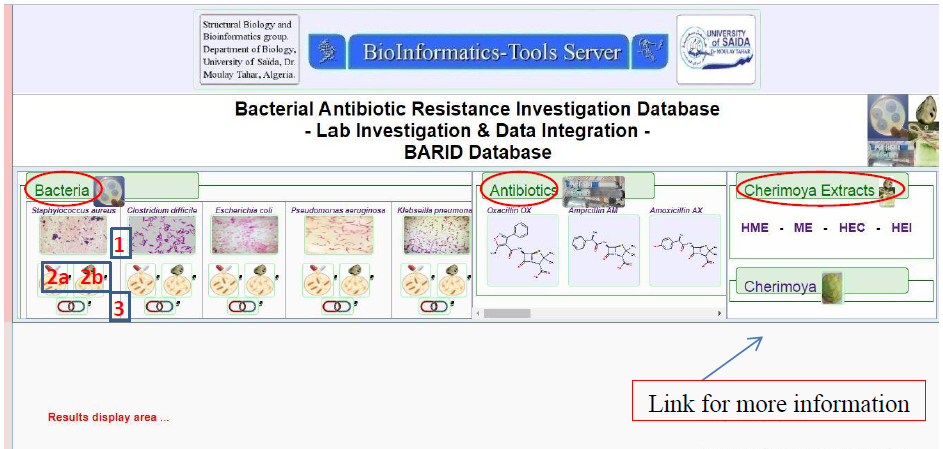
Figure4. The main interface of the BARID database system. Three ways of access/querying the database are highlighted in red; Bacteria, Antibiotics, Cherimoya (seeds) Extracts tabs. Plus an additional link “Cherimoya” to explore the Annona cherimola Mill. plant.
Querying BARID via the Bacteria-tab:
There are 4 links; denoted as 1, 2a
, 2b and 3 in (Figure 4) associated with this tab, that allow for 4 different types of results content.
1. Data results are displayed for every bacteria under study in this project. This is generated once the name or the icon image of the bacteria is clicked.
The bacteria generated data incudes its classification, principal characteristics, gram stain images. In addition, the results may contain useful data associated with proteins, genes, 3d-structures (when available), drugs information and related publications (Figure 5). This additional category of results is generated through the Data Integration Module (The system is based on an improved version of the Data Integration algorithms published in the GABagent application paper; (Abdelkrim Rachedi et al., 2000) of the BARID system.
2a. Clicking this link will generate the discovered results pertaining to the effect of all tested antibiotics on the bacteria mentioned in the Tab. This type of results helps exposing whether the bacteria, in question, show any Antibiotic Resistance against any of the antibiotics listed there in (Figure 6).
2b. This link generates the reported results related to the Anti-bacterial activity of all the four extracts on the bacteria in relation to this tab (Figure 7). Such result type exposes whether the Cherimoya seeds’ extracts have any anti-bacterial effects.
3. Through the use of this link, Joint Inspection ( ), a direct side by side inspection of antibiotics reaction and extracts effects is made possible (Figure 8). This allows direct comparison and conclusion drawing potential, refer below to the Discussion section.
), a direct side by side inspection of antibiotics reaction and extracts effects is made possible (Figure 8). This allows direct comparison and conclusion drawing potential, refer below to the Discussion section.
 ), a direct side by side inspection of antibiotics reaction and extracts effects is made possible (Figure 8). This allows direct comparison and conclusion drawing potential, refer below to the Discussion section.
), a direct side by side inspection of antibiotics reaction and extracts effects is made possible (Figure 8). This allows direct comparison and conclusion drawing potential, refer below to the Discussion section.
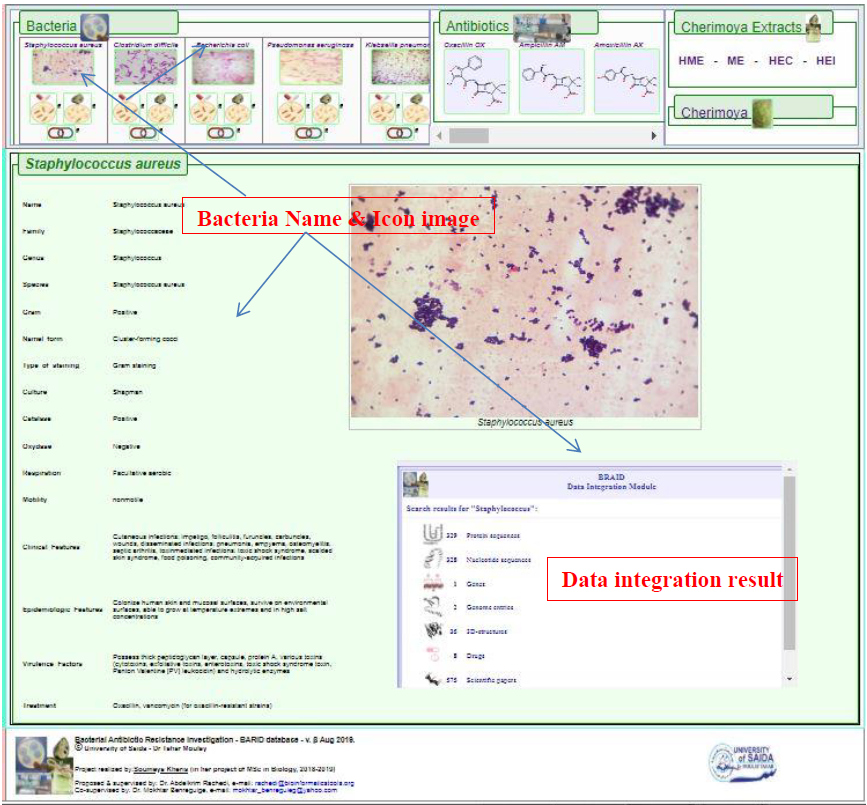
Figure5. Display of bacteria results data type after clicking the Bacteria-tab on the region marked 1 (shown in Figure 4).
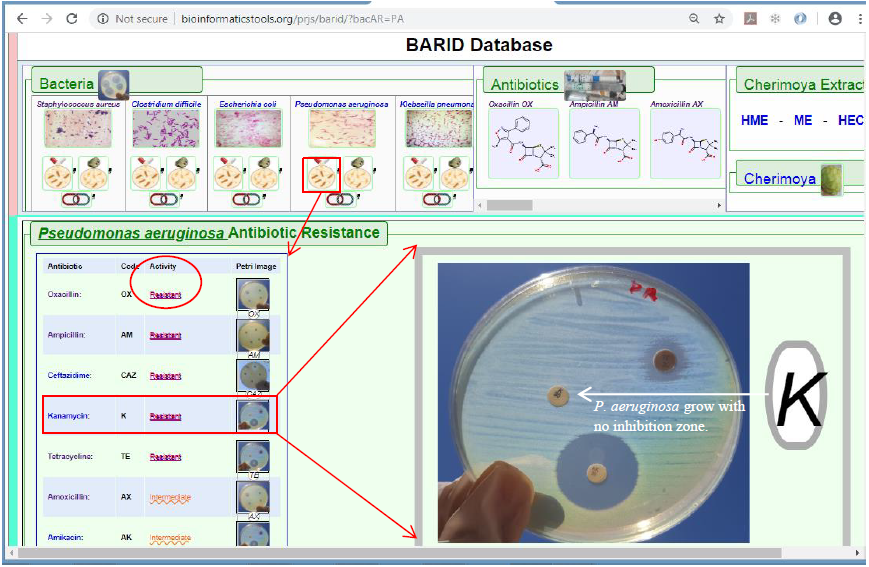
Figure6. Display of Antibiotic Resistance results data type where a summary of the bacteria reactions to all list of antibiotics used in the study is displayed. This is shown after clicking the Bacteria-tab on the region marked 2a (shown in Figure 4). Hovering with the mouse pointer over any of the petri-dish icon images will zoom in the petri-dish image for better visualization.
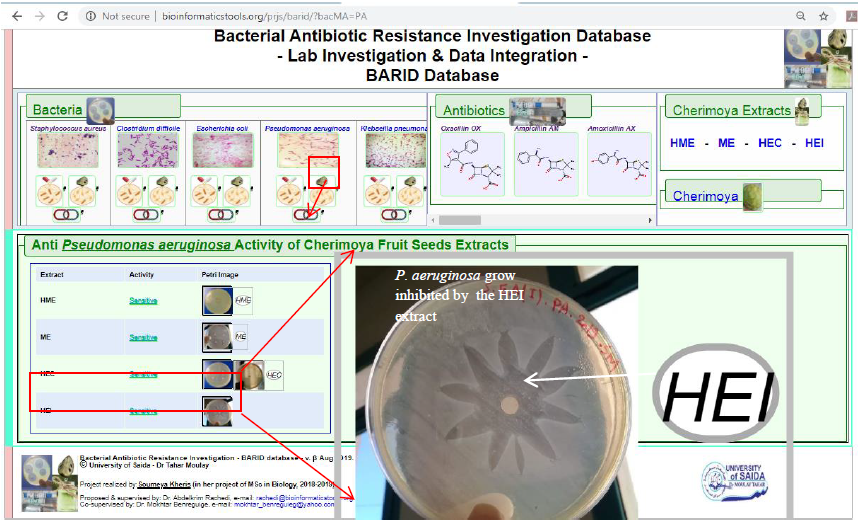
Figure7. Display of Anti-bacterial Activity results data type where a summary of the bacteria reactions to the four types of Cherimoya seeds’ extracts is displayed. This is shown after clicking the Bacteria-tab on the region marked 2b (shown in Figure 4). Hovering with themouse pointer over any of the petri-dish icon images will zoom in the petri-dish image for better visualization.
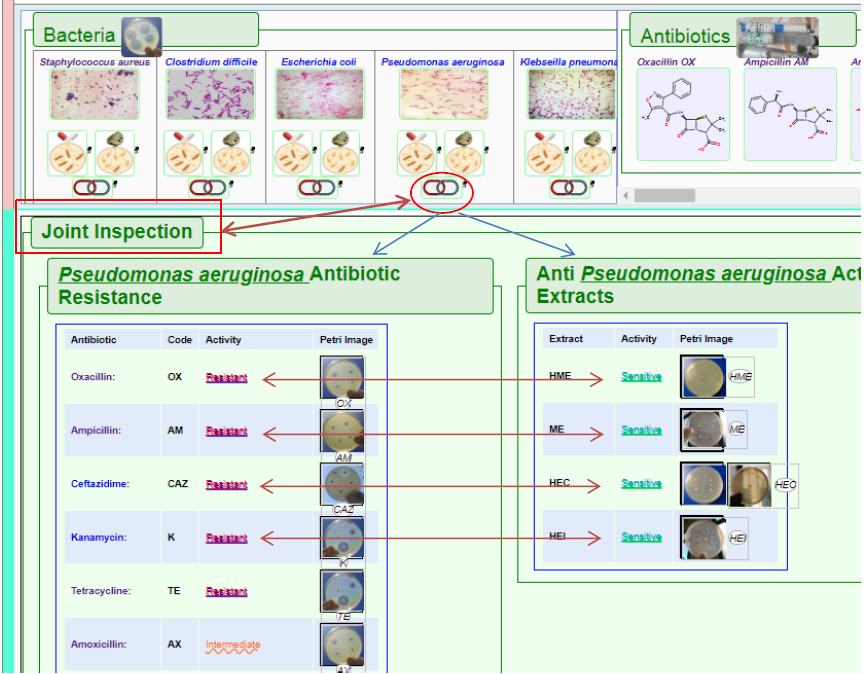
Figure8. The Joint Inspection feature
 allows for the display of side by side Antibacterial Resistance and Antibacterial effects. As seen in above figures, hovering with mouse pointer will zoom in the petri-dish icon images for better visualization.
allows for the display of side by side Antibacterial Resistance and Antibacterial effects. As seen in above figures, hovering with mouse pointer will zoom in the petri-dish icon images for better visualization.Querying BARID via the Antibiotics-tab:
Researching the database can also be done by click on the Antibiotics list provided under the Antibiotics-tab, see next figure (Figure 9).
Clicking on antibiotics by names generates summary information relevant to the antibiotic, class, type and its chemical/structure composition. Also displayed are the reactions of all of the five bacteria under study to the antibiotic in question.
Data Integration results are also generated including related proteins, genes, 3d-structures (when available), further drugs information and related publications.
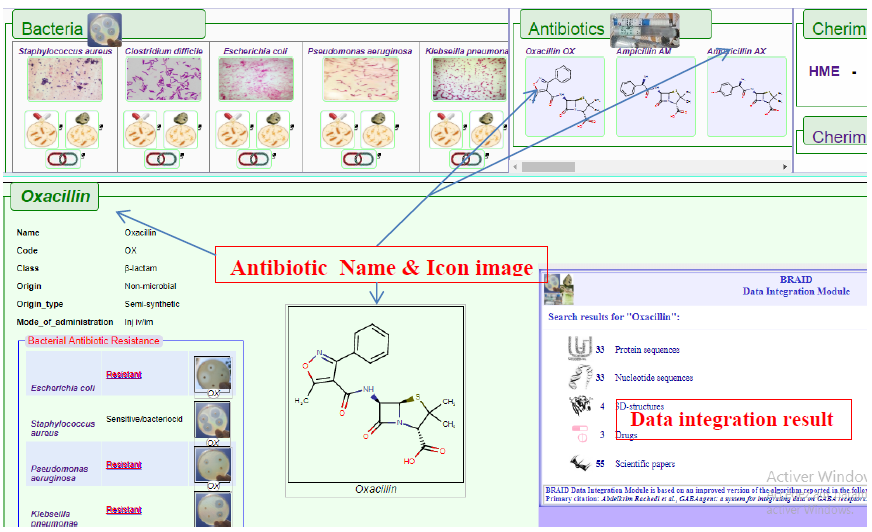
Figure9. Results shown when the Antibiotics list is used to query the database. As seen in above figures, hovering with mouse pointer will zoom in the petri-dish icon images for better visualization.
Querying BARID via the Cherimoya Extracts-tab:
This features display the anti-bacterial activity of the Cherimoya extracts against all of the five bacteria under the study as shown below in Figure 10.
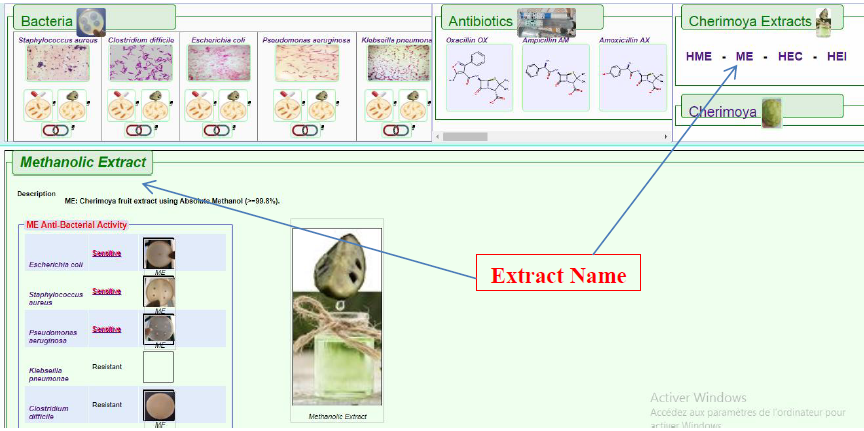
Figure10. Results shown when the Cherimoya Extracts list is used to query the database. As seen in above figures, hovering with mouse pointer will zoom in the petri-dish icon images for better visualization.
Querying BARID via the Cherimoya Fruit-tab:
This feature allows for displaying details about the Annona cherimola Mill. plant as shown below in Figure 11.
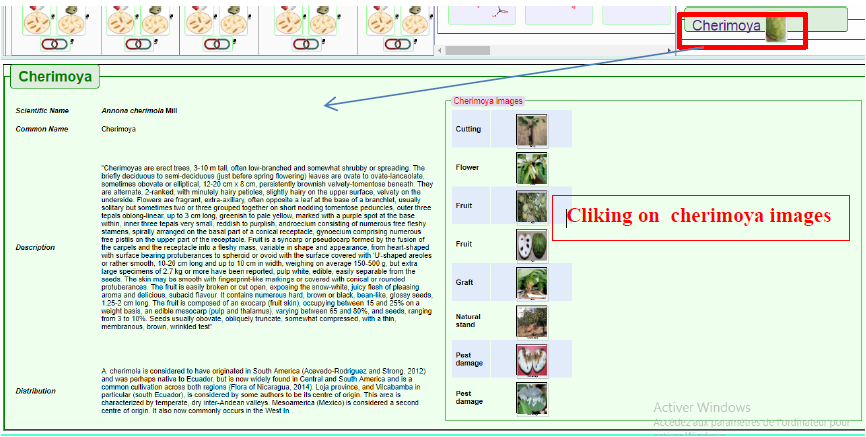
Figure11. Results shown when the Cherimoya Fruit-tab is used to query the database. Here also, hovering with mouse pointer will zoom in the petri-dish icon images for better visualization.
Data Integration:
Almost all of results mentioned above, like the shown in figures 5 and 9, would be endorsed with related useful data associated with proteins and nucleic primary sequence, genes, 3d-structures (when available), drugs information and related peer reviewed publications. This additional category of results is generated through the BARID’s Data Integration Module which is based on an improved version of the Data Integration algorithms published in the GABagent application paper; (Abdelkrim Rachedi et al., 2000).
Discussion
The discussion here concerns with the potential benefits of both venturing into such important research axes and the power of bioinformatics and its tools in exposing the scientific value found in the results data.
In addressing this subject within the context provided by the results data of obtained by the wet-lab research undertaken in this project, as represented in the BARID database, it is here preferred to take one of the virulent bacteria which is Pseudomonas aeruginosa treated as a study case and explore what the results are pointing to.
Antibiotic Resistance:
In the above case represented by Figure 6, region 2 a of the Bacteria-tab (as noted first in Figure 4) associated with Pseudomonas aeruginosa is marked by the larger red square which when clicked showed the Antibiotics Resistance of this bacteria against the list of antibiotics (only partial list is shows in the image above) pointed to by the red arrow. Hovering with the mouse pointer over any petri-dish images will zoom in the petri-dish image; in this case the zoomed in image show that P. aeruginosa is grown normal, with no zone of inhibition in the presence of the antibiotic Kanamycin (K).
This is a clear illustration that P. aeruginosa shows antibiotic resistance and is a conformation of previous published work (Mathai D et al., 2001). Same can be discovered easily in BARID about the other antibiotics and their effects on the other four bacteria.
Anti-bacterial Activity:
The case presented in Figure 7, region 2b of the Bacteria-tab (also noted first in Figure 4) associated with Pseudomonas aeruginosa, marked by the larger red square, which when clicked it provides users with the Anti-bacterial Activity of all the Cherimoya seeds’ extracts on the P. aeruginosa pointed to by the red arrow.
The zoomed in image shows a clearly visible flower-shaped zone of inhibition which is mean that P. aeruginosa is sensitive to the HEI Cherimoya seed extract type (Refer to Table 4, previous chapter).
While this study shows clear inhibition zone meaning obvious anti-bacterial activity of the extracts, results by recent study about the effect of Cherimoya extracts on (Norma et al., 2017) on P. aeruginosa show only moderate antibacterial activity.
Antibiotic Resistance and Anti-bacterial Activity:
The BARID database system provide a very simple but efficient way to compare between the results discussed above via the Joint Inspection feature (Figure 4) which brings the two sets of results together and sort them out systematically to enabling direct identification of where antibiotic ‘Resistance’ versus anti-bacterial or ‘Sensitivity’ to the Cherimoya seed extracts.
This originally novel exploration process through the BARID system is available to all of the five bacteria, antibiotics and Cherimoya extracts enabling easy and efficient investigation of the subject and help in conclusions drawing.
Conclusion
Bacterial infectious diseases represent an important cause of morbidity and mortality worldwide. An antibiotic resistant bacterium is a threat which is becoming increasingly common. The problem of microbial resistance is growing and the outlook for the use of antimicrobial drugs/antibiotics in the future is still uncertain. The search for antibacterial from natural sources has received much attention and efforts have been put in to identifying compounds that can act as suitable antibacterial /antimicrobials agent to replace synthetic ones
Screening antibacterial activity as well as for the discovery of new antibacterial compounds calls for a return to natural substances is an absolute need of our time
In this project, confirmation of ‘Antibiotics Resistance’ reported in scientific literature have been verified. In the hand high antibacterial activity of all the extracts of Annona cherimola Mill. on the virulent bacteria Pseudomonas aeruginosa is quite remarkable while also effective against the other bacteria.
Phytochemicals derived from Anonna cherimoya mill seeds serve as a prototype to develop more effective medicines in controlling the growth of bacteria .
Phytochemical screening in qualitative analysis, tests for phytochemicals were carried out and the results confirmed the presence of alkaloid. Such compounds have significant therapeutic applications against human pathogens including bacteria.
In a general conclusion, the project results clearly points to the fact that Cherimoya seeds contain a potent antimicrobial agents that need further future investigation, identification, hopefully purification and possibly chemical synthesis. Such natural products can constitute possible alternatives to antibiotics to help fight the ever growing global problem of antibiotics resistance.
This study also showed that bioinformatics and the implementation of programming and database developments are potentially essential in dealing with data generated in biology research for in managing, analysis, value adding and giving meaning to the data for maximum benefit of science and it applications in many areas of life including medicine, disease fight and biotechnology.
References
🕮 Abdelkrim Rachedi, Michael Rebhan, Hong Xue, 2000, GABAagent: a system for integrating data on GABA receptors, Bioinformatics, Volume 16, Issue 4, Pages 301–312, https://doi.org/10.1093/bioinformatics/16.4.301
🕮 Anna Perrone, Sanaz Yousefi, Alireza Salami, Alessio Papini, Federico Martinelli, 2022. Botanical, genetic, phytochemical and pharmaceutical aspects of Annona cherimola Mill, Scientia Horticulturae, Volume 296, 110896, ISSN 0304-4238, https://doi.org/10.1016/j.scienta.2022.110896
🕮 Axmark, D., & Widenius, M., 2015. MySQL 5.7 reference manual. Redwood Shores, CA: Oracle. Available online at http://dev.mysql.com/doc/refman/5.7/en/index.html
🕮 Bakken, S.S., Suraski, Z. & Schmid, E., 2000. PHP Manual: Volume 1, iUniverse, Incorporated.
🕮 D'Costa VM, Antibiotic resistance is ancient. Nature. 2011;477 (7365): 457–61. 3.
🕮 Gardiazabal F, Rosenberg G., 1993. El cultivodelchirimoyo. UniversidadCatolica de Valparaiso, Facultad de Agronomia, Valparaiso, Chile.
🕮 Microsoft Corporation, 2018. Microsoft Excel, Available at: https://office.microsoft.com/excel
🕮 National Research Council, 1989. Lost crops of the Incas: little-kown plants of the Andes with promise for worldwide cultivation. National Academy Press, Washington DC, USA.
🕮 Soumeya Kheris, 2019. Bacterial Antibiotic Resistance and Annona cherimola Mill. Seeds Antibacterial Activity Experimental Investigation & Data Annotation and Integration into an Online Accessible Database: https://bioinformaticstools.org/prjs/barid, Master thesis, Université Dr. Tahar MOULAY – Saïda Faculté des Sciences, 2018-2019, https://buc.univ-saida.dz/admin/opac_css/index.php?lvl=notice_display&id=21357
🕮 Witte W., 2004. International dissemination of antibiotic resistant strains of bacterial pathogens. Infect Genet Evol. 4(3): 187-91. 2.
