Genomic analysis of highly resistant E. faecium isolated from the gut microbiota of ampicillin treated rats.
The microbiota is the set of non-pathogenic microorganisms, bacteria, viruses, parasites and fungi, called commensals, which live in a specific environment. In humans, there are different microbiota in the skin, mouth, vagina, lungs, the intestinal microbiota is the most populated of them, hosting 1012 to 1014 microorganisms of which only a minority of bacterial species of the intestinal microbiota can be easily cultured in vitro.
This intestinal microbiota can be disturbed by the action of antibiotics during antibiotic therapy, leading to a selection phenomenon by favoring colonization by the most resistant strains (Figure 1).
There are no recommendations on methods to quantitatively measure resistance in these bacterial populations. Therefore, the assessment of resistance in a quantitative manner represents a real challenge in terms of methodologies and sampling. With this in mind, and to understand the effect of oral administration of ampicillin on the gut microbiota of rats, we developed a method to effectively quantify resistance and use enterococci as indicators to track the rate of resistance.
Results showed that treated animals excreted significantly higher percentages of resistant enterococci than the control group (P ≤ 0.05) during and after treatment, consistent with the progressive accumulation of high levels of unabsorbed ampicillin in the feces as measured by RT-HPLC. The predominant species selected after the start of treatment was Enterococcus faecium (E. faecium). Two strains of E. faecium were found to be highly resistant to ampicillin compared to the other isolates, and one showed increased resistance to the different antibiotics tested, publication link:
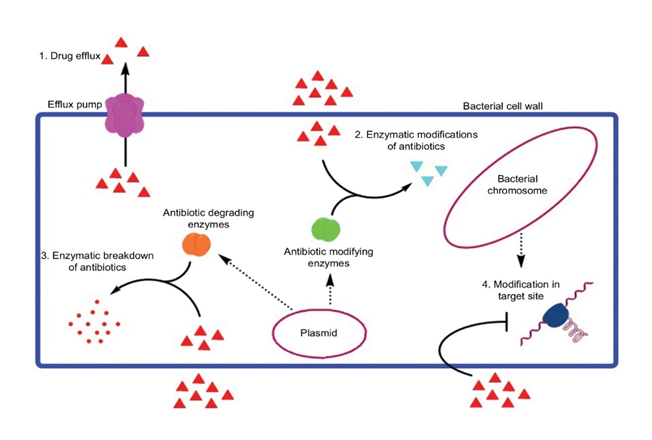
Genomic analysis of this strain indicated that its genome is composed of one chromosome and three plasmids encoding 3244 genes including 20 antibiotic resistance genes conferring different modes of resistance either by drug efflux (msrC, msrA, efmA, tetL, tetA), antibiotic-modifying enzymes (fosX, SAT4, lnu(B), AAC(6')-Ii, aph(3')-IIa, aph(3')-IIIa, ant(6)-Ia, cat), and target site modification (ponA, , dfrF, vanZ, erm(B), tetS, tetM and dfrG) (Figure 2).
Genetic mutation search showed a set of 20 mutations identified in the pbp5 gene conferring resistance to ampicillin due to an overproduction of PLP. In addition 3 virulence genes efaAfm, acm, and hylEfm were identified, 8 temperate prophages, and 2 mobile genetic elements or one is a Tn916 conjugative transposon from E. faecalis, which allows them to eventually be transferred and mobilize genes to other Gram-positive bacteria, including staphylococci, this is a part of my thesis, see the link:
Measurement of resistance within a gut microbiota is possible using enterococci as an indicator of resistance. E. faecium appears to have low pathogenicity, but may be at high risk of serving as a large reservoir of genes transmissible to other bacteria sharing the same ecosystem.
أجمل التحيات All the best