JSBB: Volume 2, Issue 2, September 2023 - AI & BIOLOGY ARTICLES
Research article: Master's research based.
Auto Protein Homology Modeling (APHM): An Educational and Research Tool for Homology Molecular Modeling.
RACHEDI Abdelkrim📧 & BAHLOUL Ouafaa
Laboratory of Biotoxicology, Pharmacognosy and biological valorisation of plants, Faculty of Natural and Life Sciences, Department of Biology, University of Saida - Dr Moulay Tahar, 20100 Saida, Algeria.
Published: 19 September 2023
Abstract
Protein structure prediction is a fundamental endeavor in molecular biology, with applications spanning from drug design to understanding cellular processes. Homology modeling, a widely used approach, leverages the structural similarity between proteins to predict the 3D structure of a target protein. In this research project, we developed a improved Auto Protein Homology Modeling (APHM), a semi-automatic tool designed to teach the principles and processes of homology molecular modeling while providing a practical platform for structure prediction. APHM integrates sequence alignment, structural superposition, and model generation in a user-friendly web interface, allowing users to predict the 3D structure of a common-core region within a protein sequence.
The structural-bioinformatics tool APHM represent a pivotal step in making structure prediction accessible and educational. It provides a hands-on experience for users to understand the principles of homology modeling. As part of its educational role, APHM has been utilised in a Masters project at the department of biology, our university where it was instrumental in generating homology models for a number of selected targets of protein sequences, enhancing the first step research and learning experiences for the students.
Availability:
Key words:
Protein structure, Protein structure prediction, Homology molecular modeling, Structural bioinformatics, Structural biology, Molecular graphics, Structure-function relationships.
Introduction
Protein structure prediction has been a longstanding challenge in molecular biology and bioinformatics, dating back to the early days of structural biology. The determination of a protein's three-dimensional structure is pivotal in understanding its function, interactions, and the underlying molecular mechanisms. Over the years, several methods and tools have been developed to address this intricate problem.
Historical Perspective
The historical roots of protein structure prediction can be traced back to the mid-20th century when the first protein structures were determined experimentally using X-ray crystallography and later, nuclear magnetic resonance (NMR) spectroscopy. Notable milestones include the determination of the structure of myoglobin by John Kendrew in 1958 (Kendrew J. C. et al., 1958) and the seminal work on hemoglobin by Max Perutz and John Kendrew in the 1960s (Perutz M. F. et al., 1960).
However, experimental structure determination is laborious, time-consuming, and often technically challenging. This limitation led to the development of computational methods for predicting protein structures. The concept of homology modeling emerged as one of the earliest strategies, where the structure of a target protein is inferred based on the known structure of a homologous protein. This concept laid the foundation for modern homology modeling approaches like the one implemented in Auto Protein Homology Modeling (APHM).
The Evolution of APHM
The origins of Auto Protein Homology Modeling (APHM) can be traced back to its original development during a Ph.D. thesis in 1994 (Rachedi A., 1994). The thesis, titled "Three-dimensional structural studies on dihydrofolate reductase," marked the inception of APHM as a structural-bioinformatics tool for exploring protein structures solved during the Ph.D. project. Over the years, APHM has undergone continuous refinement and improvement, culminating in the version subject to this article and the Masters thesis project subject to implementing it (Bahloul, O., 2022).
The version of APHM presented here represents an advanced iteration of the original tool, integrating state-of-the-art methodologies, user-friendly interfaces, and enhanced performance. This evolution reflects the ongoing commitment to advancing structural biology tools to meet the growing demands of researchers and educators in the field.
Current State of Structure Prediction
In the present era, protein structure prediction has evolved into a multidisciplinary field encompassing bioinformatics, computational biology, and structural biology. Several computational methods have been developed, including ab initio modeling, threading, and molecular dynamics simulations. Each method has its own strengths and limitations, making them suitable for different scenarios.
Despite significant advancements, protein structure prediction remains a complex challenge due to the vast conformational space that proteins can explore. The accuracy of predictions varies depending on factors such as sequence similarity, template availability, and the sophistication of the modeling algorithm. Consequently, the field continually seeks innovative solutions to improve the accuracy and efficiency of structure prediction.
Implementations and Challenges
Tools such as APHM play a crucial role in democratising structure prediction and making it an educational endeavor. They offer users a tactile learning experience, allowing them to grasp the fundamental principles of homology modeling. As a testament to its educational value, APHM has been actively employed in a Masters project titled 'Molecular Modeling and Prediction of Protein 3D Structure: Principles and Applications' (Bahloul, O., 2022). In this project, APHM was instrumental in generating models for various selected target sequences, enriching students first research steps and their educational journey.
Moreover, in serving the research aspect, APHM is made available online for structure prediction experiments related to understanding structure-function relationships and investigating ligand binding sites. This extends its utility beyond the educational realm to support research endeavors, facilitating investigations into the molecular underpinnings of various biological processes.
However, it is important to acknowledge that this improved version of APHM offers valuable insights, hands-on training and education in the field of structural biology, it is still not aimed at high-precision modeling in research applications. Nevertheless, it provides an essential platform for bridging the gap between theory and practice in the fascinating field of protein structure prediction.
Methods
Sequence Alignment and Template Selection
The first step in homology modeling is identifying homologous protein sequences with known structures. To achieve this, APHM utilises the Basic Local Alignment Search Tool (BLAST) tool (Altschul S.F. et al., 1990) to align the target protein sequence against a comprehensive Protein Data Bank (PDB) based sequence library (Berman H.M. et al., 2000). Highly similar sequences with known structures are selected as potential templates.
This pivotal step involves identifying homologous protein sequences with known structures that closely resemble the target protein of interest. In APHM, template selection is a meticulous process driven by rigorous criteria such as high sequence identity, low E (expectancy) values, and robust alignment scores, all of which contribute to the reliability of the subsequent modeling process.
Incorporating these stringent criteria into the template selection process safeguards the precision and reliability of the subsequent homology modeling endeavor. By choosing highly similar sequences with known structures that pass the rigorous tests of sequence identity, low E values, and high alignment scores, APHM enhances the likelihood of generating an accurate 3D model.
The sequence alignment and template selection constitute the foundational steps of homology modeling within APHM. This process is not a mere selection of templates; it is a meticulous curation of templates that meet exacting standards, setting the stage for the creation of structurally and functionally meaningful 3D-protein models.
Structural Superposition: Finding the Optimal Template
After identifying potential templates, the next critical step is structural superposition. APHM harnesses the power of online tools to execute this crucial task (Anfinsen C. B., 1973). Structural superposition involves aligning the 3D structures of the selected template proteins seeking the best fit. The aim is to identify the template structure that exhibits the lowest Root Mean Square Deviation (RMSD), signifying the closest structural resemblance to the target (Sali, A., & Blundell, T. L., 1993).
The RMSD is a key metric used to quantify the overall dissimilarity between two protein structures. A lower RMSD indicates a higher level of structural similarity between the target and template, a pivotal factor in ensuring the reliability of the subsequent modeling process. The template structure with the lowest RMSD is chosen as the foundation for generating the 3D model in APHM.
Common-Core Structure Identification: Unearthing the Shared Foundation
Structural superposition not only serves the purpose of template selection but also holds the key to identifying common-core structure regions shared among the selected PDB structures. These common-core regions represent the structurally conserved segments within the target protein and the chosen templates (Anfinsen C. B., 1973). The common-core region serves as the foundational blueprint for generating the 3D-model within APHM process that leads to the identification of structurally conserved elements across multiple templates and ensures that the resulting model captures the essential structural features critical for the protein's function and interactions.
The identification of common-core regions highlights the conserved structural elements, enabling APHM to create 3D models that capture the essential features shared across related proteins. This not only aids in understanding the structural basis of function but also provides a solid foundation for further research and experimentation. These steps ensure that the selected template closely aligns with the target and that the resulting 3D model encapsulates the vital structural elements, setting the stage for meaningful insights into protein function and interactions.
APHM's Workflow
APHM's user-friendly interface facilitates the modeling process and is accessible online from the following link: https://bioinformatics.univ-saida.dz/bit2/?arg=MD2. Users input the target protein sequence, specify the template protein structure by providing its PDB identifier, and highlight the common-core region segments. Upon clicking the "Submit" button, APHM generates the predicted 3D model of the common-core region in PDB format, preserving the correct amino acid substitutions.
Common-Core Visualisation
One of the distinguishing features of APHM is its ability to provide users with an interactive and dynamic visualisation of the predicted common-core structure. After APHM generates the 3D model of the common-core region, users are presented with an option that initiates a visualisation window. This window utilises the Mol* (Molstar) API, a powerful molecular graphics system (Sehnal D. et al. 2018, Sehnal D. et al. 2021), to display the 3D-structure. This dynamic visualisation component adds a valuable dimension to the APHM tool, empowering users to explore and appreciate the intricacies of protein structures.
Example Usage of APHM
To illustrate the practical application of Auto Protein Homology Modeling (APHM), let's consider an example in which we predict the 3D structure of the human Dihydrofolate Reductase (DHFR) protein. We have extracted the target protein sequence with the accession code P00374 from the UniProt database (UniProt Consortium 2019), Figure 1.
Figure 1. The amino-acids sequence of the human DHFR in FASTA format (Pearson, W. R., & Lipman, D. J. 1988) was downloaded from the UniProt database as explained in text above. The first line after the symbol “>” is a definition line that provides relevant information about the sequence. The remaining lines are the amino-acids sequence represented in the single letter code of amino-acids.
In this example, the Protein Data Bank (PDB) structure with the identifier 1U70 is used as the template. The common-core region we have selected for modeling includes residues: 4 – 12, 14 – 22, 26 – 38, 48 – 60, 66 – 77, 88 – 91, 112 – 125, 131 – 138, 145 – 149, and 175 – 184.
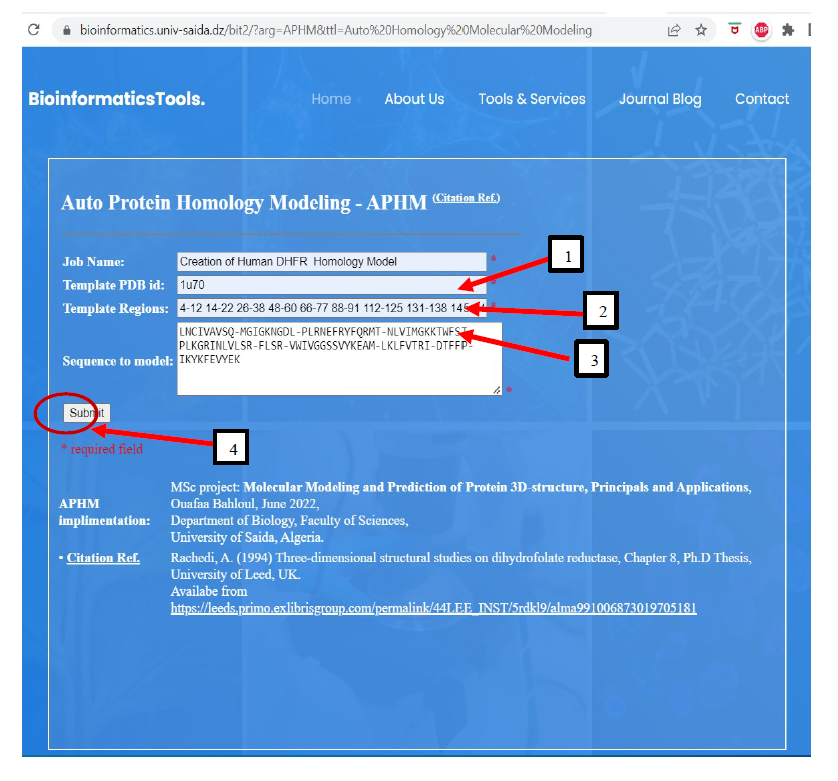
To begin the modeling process, the user accesses APHM's user-friendly interface. Below is a visual representation of the APHM interface form, highlighting the input data for this example, Figure 2:
To begin the modeling process, the user accesses APHM's user-friendly interface. Below is a visual representation of the APHM interface form, highlighting the input data for this example, Figure 2:
Figure 2. APHM interface with input data of structure template 1U70 1,
core regions 2
and amino-acids sequence 3. To build the side-chain and model, the user need to click
button Submit encircled in red 4.
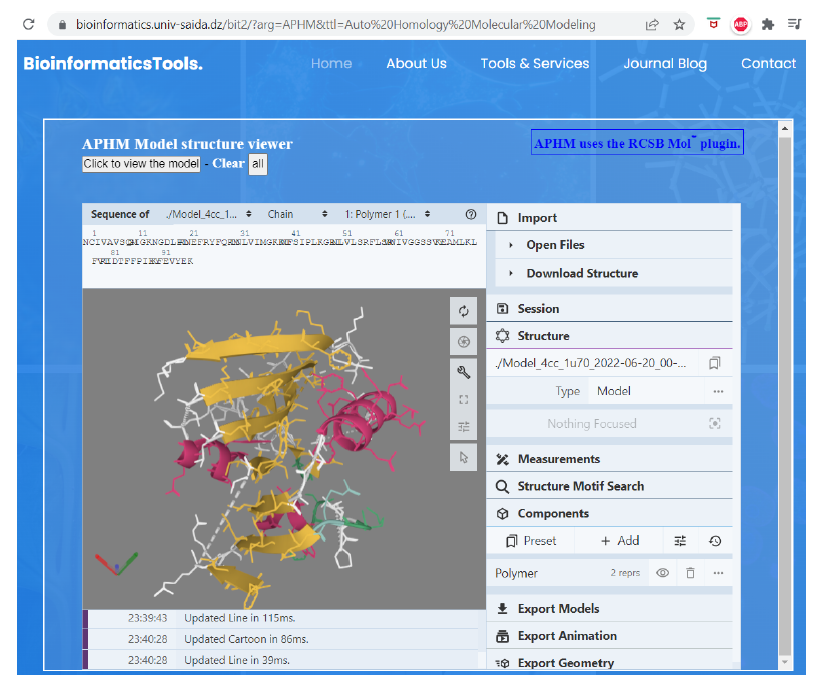
Upon clicking the "Submit" button, see Figure 1 above, APHM processes this input data to generate the predicted 3D model of the common-core region, preserving the correct amino acid substitutions, Figure 3.
Figure 3. The APHM 3D-viewer displaying the predicted model created based on the DHFR sequence shown in Figure 1 and core regions suitable to the template structure 1U70.
This example showcases how APHM empowers users to perform homology modeling with ease and precision, making it a valuable tool for both educational and research purposes
Limitations
It is important to note that APHM has certain limitations. It does not generate loop regions, and it does not apply energy minimization to the final 3D structure model. However, these limitations are intentional, as APHM's primary purpose is educational, allowing users to grasp the fundamentals of homology modeling.
Results and Discussion
APHM was developed with a focus on educational purposes, making it a valuable resource for teaching and learning about homology modeling. Its user-friendly interface simplifies the often complex process of structure prediction, allowing students and researchers to gain hands-on experience.
One of the prominent outcomes of APHM's educational application is the empowerment of learners to actively engage with the principles of homology modeling. By aligning target sequences with known structures, selecting suitable templates, and highlighting common-core regions, users gain profound insights into the critical factors influencing structural predictions. This hands-on learning experience not only demystifies the complex world of structural biology but also nurtures a generation of scientists adept at applying these techniques.
Furthermore, the common-core structure visualization component, powered by the Mol* API (Sehnal D. et al. 2018, Sehnal D. et al. 2021) facilitates a dynamic exploration of protein structures. Users can rotate, zoom, and manipulate the visual representation, fostering an enhanced understanding of the spatial arrangements of amino acids. This dynamic interaction enriches the educational experience, making abstract concepts tangible and comprehensible.
While its educational utility is evident, APHM also holds significant promise in the realm of advanced research. The ability to generate 3D models with precision, driven by stringent template selection criteria and structural superposition, equips researchers with a versatile tool for exploring diverse biological questions.
APHM is poised to play a pivotal role in unraveling structure-function relationships within proteins. The fidelity of its models, rooted in rigorous template selection and common-core structure identification (Anfinsen C. B., 1973), enables researchers to probe the structural basis of protein function. Whether investigating enzyme catalysis, ligand binding sites, or allosteric regulation, APHM offers a platform for in-depth exploration.
In this simple case of predicting the structure of enzymes such as the DHFR, the common-core region of the generated model would contain ligand binding sites which may bind substrates and cofactors. The model thus can be used in understanding how ligand binding happens and model the effects of amino-acids substitutions (mutations) that may exist in the binding sites. Such type of insights contributes directly into structure-function relationship in proteins.
As the field of structural biology continues to advance, there is a growing need for integrating diverse sources of data, such as protein-protein interaction networks, co-evolutionary information, and advanced machine learning techniques, into structure prediction methods. These approaches have the potential to enhance accuracy and extend the applicability of structure prediction tools in understanding complex biological processes.
Conclusion
Auto Protein Homology Modeling (APHM) serves as an innovative tool for both educational and research purposes. Its simplified yet informative approach to homology modeling empowers users to understand the core principles of this critical field. While the approach is not high-precision modeling one, APHM provides a solid foundation for students and young researchers interested in exploring the fascinating world of protein structure prediction.
The successful implementation of APHM in a Masters project (Bahloul, O., 2022), have demonstrated its potential capabilities in both sectors of education and research in the realm of structure prediction and modelling:
As an educational tool, APHM demystifies the complexities of homology modeling, nurturing a generation of scientists equipped with the skills to navigate structural biology's intricacies. Through hands-on learning and dynamic visualization, it fosters a profound understanding of the principles underlying protein structure prediction. In the research arena, APHM stands as a versatile instrument, offering precision and reliability in modeling. Researchers can harness its capabilities to explore structure-function relationships, investigate ligand binding sites, and embark on explorations that unveil the mysteries of protein biology.
As an educational tool, APHM demystifies the complexities of homology modeling, nurturing a generation of scientists equipped with the skills to navigate structural biology's intricacies. Through hands-on learning and dynamic visualization, it fosters a profound understanding of the principles underlying protein structure prediction. In the research arena, APHM stands as a versatile instrument, offering precision and reliability in modeling. Researchers can harness its capabilities to explore structure-function relationships, investigate ligand binding sites, and embark on explorations that unveil the mysteries of protein biology.
The journey of APHM, from its inception during a Ph.D. thesis in 1994 to its current advanced iteration, signifies its enduring relevance and adaptability. APHM's evolution reflects the commitment to advancing structural biology tools to meet the ever-evolving demands of researchers and educators in the field.
References
🕮 Kendrew, J. C., et al. (1958). A three-dimensional model of the myoglobin molecule obtained by X-ray analysis. Nature, 181(4610), 662-666.
🕮 Perutz, M. F., et al. (1960). Structure of haemoglobin: A three-dimensional Fourier synthesis at 5.5-Å. Resolution, obtained by X-ray analysis. Nature, 185(4711), 416-422.
🕮 Rachedi, A. (1994) Three-dimensional structural studies on dihydrofolate reductase, Chapter 8, Ph.D Thesis, University of Leed, UK. https://leeds.primo.exlibrisgroup.com/permalink/44LEE_INST/5rdkl9/alma991006873019705181
🕮 Bahloul, O. (2022). Molecular Modeling and Prediction of Protein 3D-structure, Principals and Applications, Master Thesis, University of Saida, Algeria, June session.
🕮 Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. (1990). NCBI BLAST: a better web interface. Nucleic Acids Research. J. Mol. Biol. Oct 5, 215(3), 403-410 Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. (2000). The Protein Data Bank. Nucleic Acids Res. 28:235–242.
🕮 Anfinsen, C. B. (1973). Principles that govern the folding of protein chains. Science, 181(4096), 223-230.
🕮 Sehnal D., Rose A.S., Koča J., Burley S.K., Velankar S.. Byška J., Krone M., Sommer B.. (2018). Mol*: Towards a common library and tools for web molecular graphics. Proceedings of the Workshop on Molecular Graphics and Visual Analysis of Molecular Data. 29–33.
🕮 Sehnal D., Bittrich S., Deshpande M., Svobodová R., Berka K., Bazgier V., Velankar S., Burley S.K., Koča, J. and Rose A.S. (2021). Mol* Viewer: modern web app for 3D visualization and analysis of large biomolecular structures. Nucleic Acids Res. Jul 2; 49(W1): W431–W437.
🕮 UniProt Consortium. (2019). UniProt: a worldwide hub of protein knowledge. Nucleic Acids Research, 47(D1), D506-D515.
🕮 Pearson, W. R., & Lipman, D. J. (1988). Improved tools for biological sequence comparison. Proceedings of the National Academy of Sciences of the United States of America, 85(8), 2444-2448.
🕮 Sali, A., & Blundell, T. L. (1993). Comparative protein modelling by satisfaction of spatial restraints. Journal of Molecular Biology, 234(3), 779-815.