Genomes and Mutations in the currently circulating variants of Omicron virus.
Find variants data in the SVGM-Explorer 
Abdelkrim RACHEDI
Laboratory of Biotoxicology, Pharmacognosy and biological valorisation of plants, Faculty of Sciences, Department of Biology, University of Saida - Dr Tahar Moulay, 20100 Saida, Algeria.
📧 E. mail: abdelkrim.rachedi@univ-saida.dz
Published: 08 December 2022
In the light of the current status of COVID-19 around the world which though it may have subsided and
gone to a lower profile, in many region like China, USA, France, UK, to mention few, the new SASR-Cov-2 variants still posing tough challenges to health.
Several surges of new Omicron (B.1.1.529) subvariants have showed up over the late few months including BA.1, BA.2, BA.3, BA.4, BA.5, BA.2.75, BA.2.12.1 and XE subvariants. More importanly drawing
attention are the subvariants of BA.5 namely the BQ.1 and BQ.1.1 all of which are now constituing a large percentage
of SARS-CoV-2 infection reaching over 60% in the USA.
The SVGM-Explorer (SVGM-exp β-v.1.0)⭕ is a bioinformatics tool module part of the VirusesDb β-v.4.0🔶 database, see the link below,
presents data in a "comparison-fashion" to make it easy for the users (scientists/students) to spot differences in the mutations found in the different
variants.
The full genomic sequences of the currently circulating variants of Omicron BA.1, BA.2, BA.3, BA.4, BA.5, BA.2.75
and BA.2.12.1, Figure 1, are available via the SVGM-Explorer link below:

Figure 1. The SVGM-Explorer comparision-fashion table presentaion of SARS-CoV-2 variants including Omicron ones.
In order to explore the genome sequence and mutations of any of the variants, users need to click on an accession-code of a relevant variant, for example,
the code OP022774 which is one of the BA.4 omicron variants. Clicking on the such accession-code, trigers SVGM-Explorer to display a window that enbles exploring and downloading
the genomic sequence and over novel 30 mutations (compared to previous variants), see Figures 2. and 3.
The results can also be generated using the below SVGM-Explorer link which can be composed for any other variants in shown in the list by substituting the accession-code value (id value in the link):
The results can also be generated using the below SVGM-Explorer link which can be composed for any other variants in shown in the list by substituting the accession-code value (id value in the link):
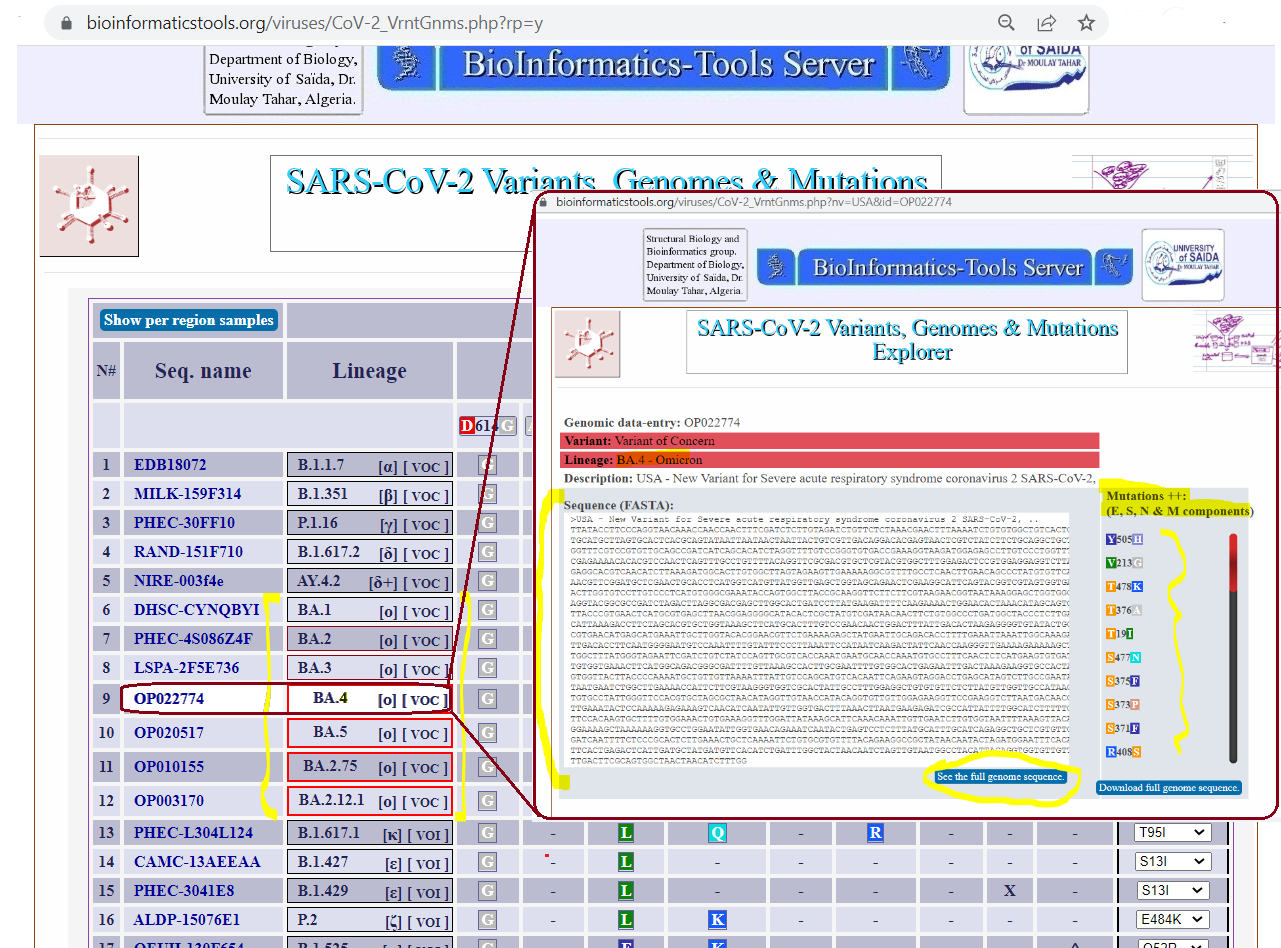
Figure 2. SVGM-Explorer demonstration show of genomic sequence and mutations once variants accession codes are clicked.
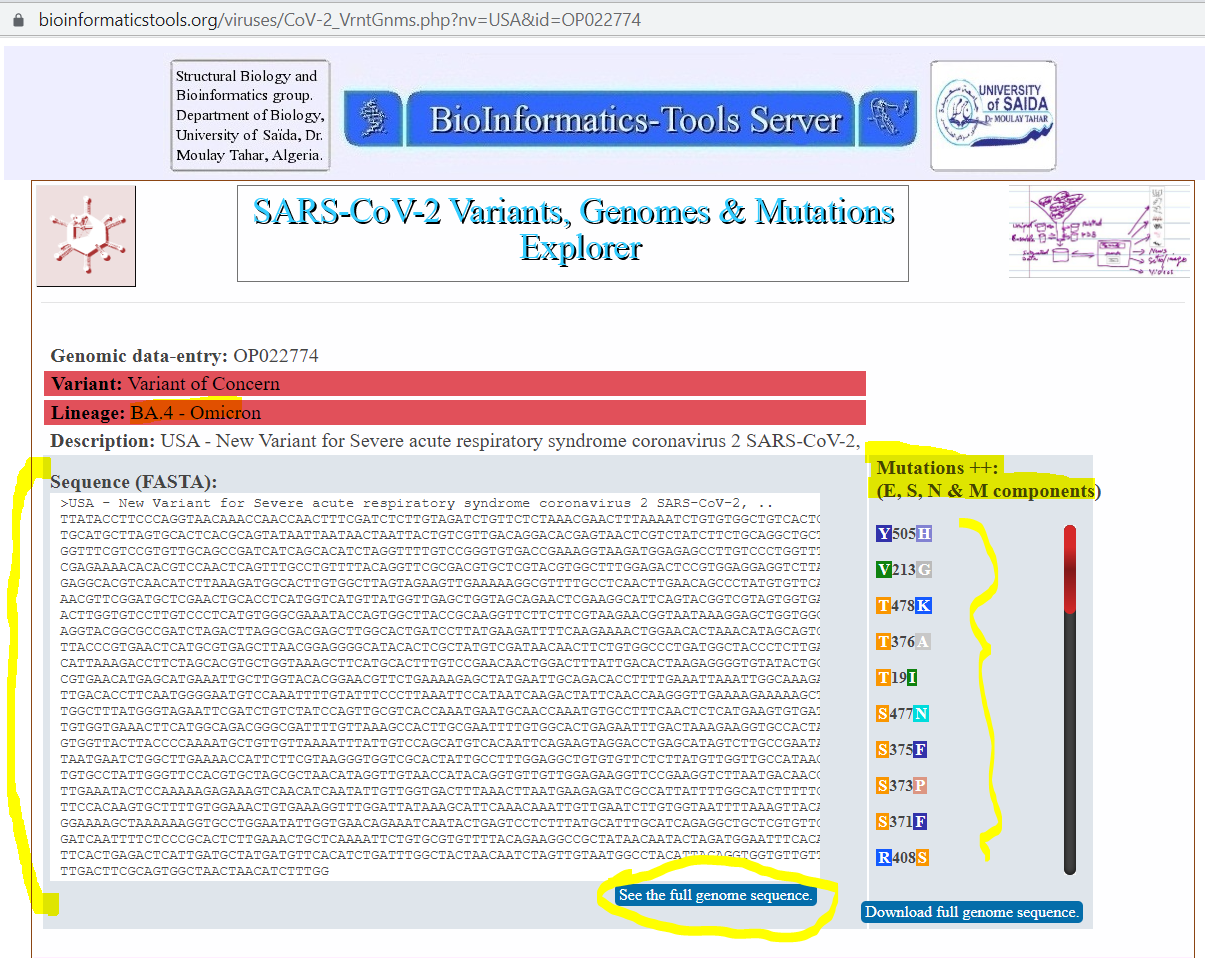
Figure 3. Details of genomic sequence and mustations for the Omicron variant BA.4 associated with accession-code OP022774
In this regards, it would be worthwhile to realise that the ability to spot differences in the mutations and their types between the existing
and emergin variants help greatly in assessment of the ability of the available vaccines to provide immunity against the new strains of the COVID-19 disease.
Would also help in understanding their potential of disease causing and come up with proper strategies for limiting its spread between people and communities and in principal acquiring the visions to design new drugs and inhibitor medicines and cures against the running unfortunate pandemic disease.
While most the new Omicron variants, including others curently not available in SVGM-Explorer, showed faster propagation in infection, health authorities around the world have not found evidence of more severe illness caused by the variant.
However, a number of the new mutations, in Omicron variants, have demonstrated escaping immunity from previous infection and vaccination.
In relation to the subject of genomic data and mutations data integration and developement at the University of SAIDA, Algeria, refer to the JSBB articles under the themes Genomics & COVID-19 and Bio-Databses, the links below:
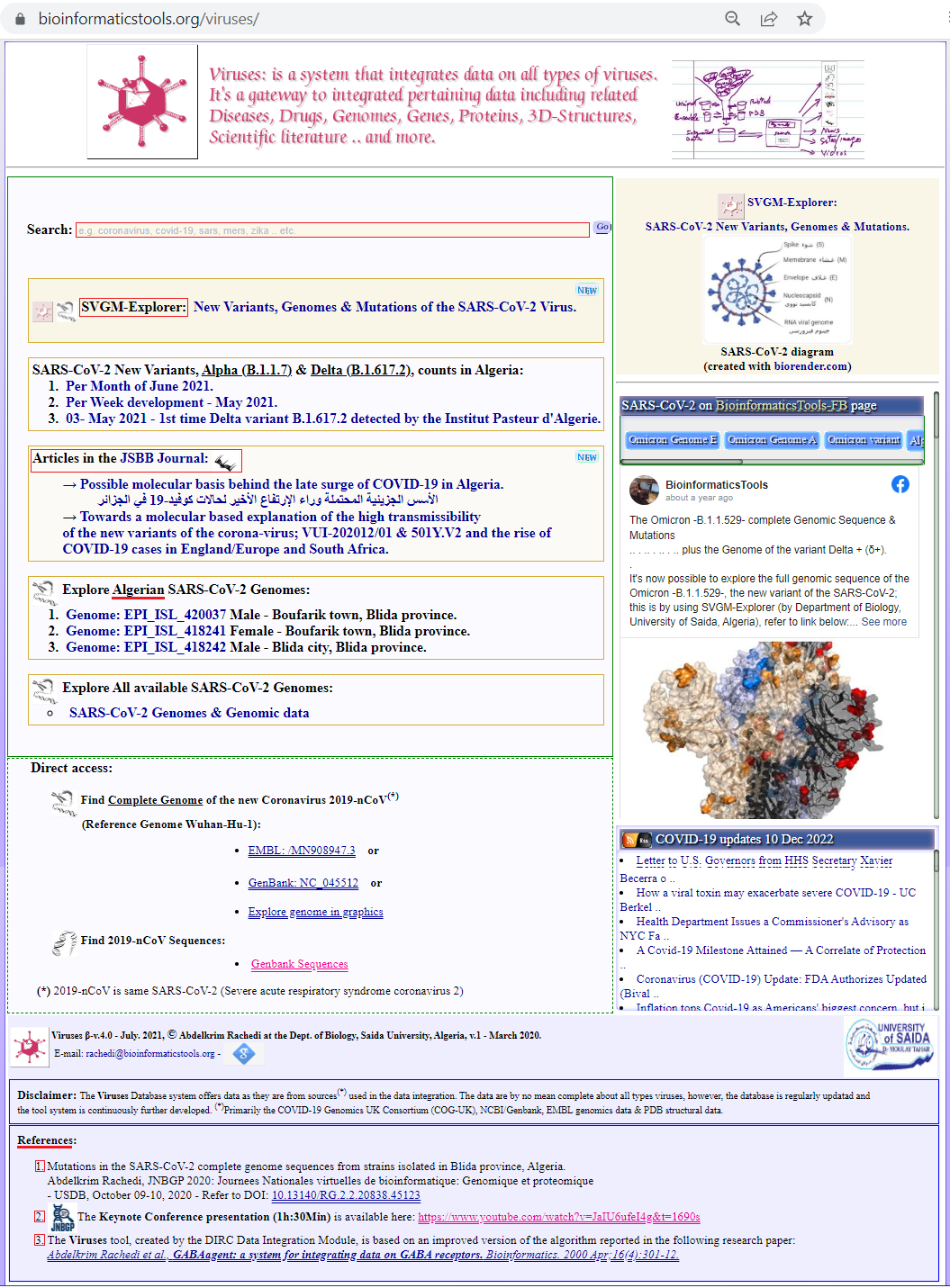
See also the JSBB article about the VirusesDb🔶, see Figure 4 below:
🖝 VirusesDB a new tool to finding scientific data on 2019-nCoV.
تطبيق VirusesDB مصدر للبيانات العلمية المدمجة حول فيروس كورونا المستجد.

Figure 4. The VirusesDb database interface page.
For more about the Omicron variants, SARS-CoV-2 virus and COVID-19 progress, refer to following links:
Figures
References for the SVGM-exp β-v.1.0 & VirusesDb β-v.4.0:
- Mutations in the SARS-CoV-2 complete genome sequences from strains isolated in Blida province, Algeria.
Abdelkrim Rachedi, JNBGP 2020: Journees Nationales virtuelles de bioinformatique: Genomique et proteomique
- USDB, October 09-10, 2020 - Refer to DOI: https://dx.doi.org/10.13140/RG.2.2.20838.45123  The Keynote Conference
presentation (1h:30Min) is available here: https://www.youtube.com/watch?v=JaIU6ufeI4g&t=1690s
The Keynote Conference
presentation (1h:30Min) is available here: https://www.youtube.com/watch?v=JaIU6ufeI4g&t=1690s
- The VirusesDb, data annotation & integration tool, created by the DIRC Data Integration Module, is based on an improved version of the algorithm reported in the following research
paper:
Abdelkrim Rachedi et al., GABAagent: a system for integrating data on GABA receptors. Bioinformatics. 2000 Apr;16(4):301-12.
⭕ SVGM-exp β-v.1.0: SARS-CoV-2 Variants, Genomes %26 Mutations Explorer, by Structural Biology & Bioinformatics Groups, Biology Dept, Faculty of Science, University of Saida, Algeria
نظام: مستكشف متغيرات و طفرات فيروس SARS-CoV-2 ، فريق البيولوجيا ثلاثية
الأبعاد و المعلوماتية الحيوية، قسم البيولوجيا، كلية العلوم، جامعة سعيدة، الجزائر
🔶 VirusesDb β-v.4.0: Viruses Genomes & Data-ntegration Database, by Structural Biology & Bioinformatics Groups, Biology Dept, Faculty of Science, University of Saida, Algeria
نظام: قاعدة بيانات VirusesDb لجينومات الفيروسات و بيانات مدمجة ، فريق البيولوجيا ثلاثية
الأبعاد و المعلوماتية الحيوية، قسم البيولوجيا، كلية العلوم، جامعة سعيدة، الجزائر