JSBB: Volume 3, Issue 4, February 2025 -
ANTIBIOTICS RESISTANCE
Research article – Review type.
Systematic review of genetic determinant and prevalence of resistance in bovine mastitis pathogens in Algeria
BOUNOUA Cheimaa📧 & SOUNA Djahida
Department of Biology, Laboratory of Molecular Biology, Genomics and Bioinformatics, Faculty of Nature and Life Sciences, University Hassiba Benbouali of Chlef, Algeria.
📧 BOUNOUA Cheimaa - E. mail: c.bounoua@univ-chlef.dz
Published: 10 February 2025
Abstract
Mastitis is one of the most problematic and costly production diseases of dairy cattle. It is frequently treated
with broad-spectrum antimicrobials. The aim of this paper is to document the current situation of antimicrobial
resistance identified in the main pathogens causing bovine mastitis in Algeria. Genetic determinants and prevalence
of resistance to different antimicrobials are discussed. Only studies focusing on bovine mastitic milk samples that
investigated antibiotic-resistant bacteria were selected and just 7 articles met the inclusion criteria and were
included in this systematic review. Data were narratively synthesized and analyzed.
The most commonly isolated bacteria were Staphylococcus aureus and Escherichia coli. The findings indicate
signs of antibiotic misuse in Algeria, coupled with the rapid dissemination of resistant bacterial strains and
inadequate surveillance, contributing to the problem. The resistance profile of the agents studied in Algeria showed
alarmingly high levels of resistance to widely used antimicrobial drugs for the treatment of mastitis. Several genetic
determinants were detected (bla CTX-M, tetA, bla TEM-1, mecA, tetM, blaZ).
Without critical measures to stop the excessive use of antibiotics, the prevalence of antibiotic-resistant bacteria
could rise posing major risks to both animal and human health. Therefore, routine antimicrobial susceptibility
testing before treatment is crucial for selecting effective antibiotics and preventing resistance development.
Keywords: Antibiotic therapy, Bovine mastitis, Resistance genes, Algeria, Systematic review
Introduction
Mastitis is one of the most prevalent diseases in dairy cows, which has an impact on the udder's health as well as the quality of milk yielded (Tommasoni et al., 2023).Treatment for mastitis often consists of using broad-spectrum antibiotics, which are known to partially enhance resistance evolution, without any prior knowledge of cause-related agents. This increases the selective pressure on potentially present pathogens and is considered a potential human health risk (Rana et al., 2022). Methicillin-resistant Staphylococcus aureus (MRSA) and multidrug resistant Gram-negative bacteria are considered a key challenge in mastitis treatment (Naranjo-Lucena and Slowey, 2023).
Mastitis control requires a comprehensive strategy, including herd disease prevention, antimicrobial stewardship, and targeted antibiotic therapy based on pathogen identification and resistance patterns. This is because there are many subclinical infections, withdrawal periods, and antimicrobial resistance (Ruegg, 2021).
Using the "One Health Approach" is essential to addressing the challenges caused by antibiotic resistance in the management of mastitis. This approach recognizes the interdependency of human, animal, and environmental health, striving to enhance comprehension and control of antimicrobial resistance concerning mastitis and other food. Key steps in this approach involve monitoring the prevalence of antimicrobial resistance in mastitis pathogens and implementing effective control measures (Sipahi et al., 2023).
Thus, this review aims to document the current situation of antimicrobial resistance identified in the main pathogens causing bovine mastitis in Algeria. Genetic determinants and prevalence of resistance to different antimicrobials are discussed.
Methodology
An exhaustive search was undertaken across various databases, including Google Scholar, PubMed, Scopus and ScienceDirect, to identify relevant studies published over time. The search strategy incorporated specific headings and keywords such as "Antibiotic therapy," "bovine mastitis," "Algeria," and "Resistance genes." The review methodology involved a meticulous analysis of each identified paper, with a focus on extracting pertinent information from all sections to compile a comprehensive synthesis. To ensure data integrity, duplicate papers were systematically removed, and the remaining data underwent rigorous screening. Irrelevant works were excluded, and full-text documents were subsequently screened. The inclusion criteria were defined to encompass articles conducted in Algeria that specifically addressed antibiotics resistance and resistance genes isolated from agents responsible for bovine mastitis. Exclusion criteria were applied to studies that did not utilize bovine milk mastitis samples, did not discuss resistance genes, or were not conducted in Algeria, Table 1.
Results
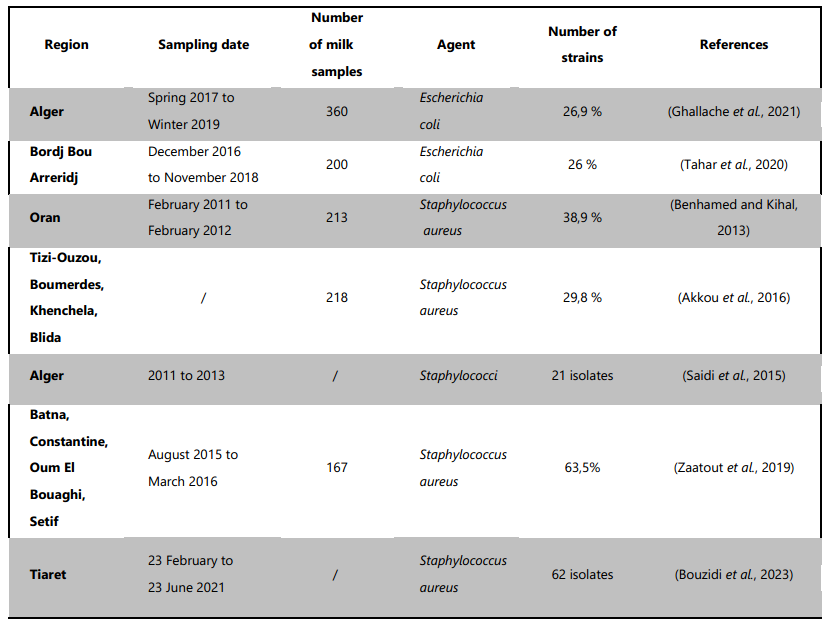
The systematic review identified a limited pool of literature meeting our predefined inclusion criteria. Only seven articles were deemed eligible for inclusion in our analysis. It is important to note that these studies were conducted in different parts of Algeria and at different periods and they used different methods to study and analyse. Even so, the prevalence of the bacteria may have changed over time and results may not be directly comparable. All of the eligible studies described the occurrence of antimicrobial resistance while all the studies did not provide information on farms types. The details of the studies regarding the countries; author name; year; Sampling date; reported prevalence of bacteria are given in Table 1.
Table 1. Prevalence of S. aureus and E. coli in Algerian dairy cows, highlighting a high occurrence of both pathogens, with significant antibiotic resistance patterns observed.
Discussion
Mastitis can be categorized into clinical and subclinical mastitis based on different features. Clinical mastitis is characterized by visible abnormalities in the milk, such as flakes, clots, or watery secretions, as well as swollen, hot, and painful quarters. In acute cases, general signs like hyperthermia, anorexia, and depression can also be present (Cobirka et al., 2020). On the other hand, subclinical mastitis is not easily diagnosed due to the lack of evident signs in the milk or the affected animals. Its main symptoms are associated with an increased somatic cell count and decreased milk production (Cobirka et al., 2020).
Subclinical mastitis, as evidenced by studies in Algeria has been shown to have a high prevalence, with rates of 66.4% (Ghallache et al., 2021), 52.12% (Benhamed and Kihal, 2013b) and 40% (Zaatout et al., 2019) based on California Mastitis Test (CMT) results. This is concerning due to the potential for a longer duration of subclinical mastitis compared to clinical mastitis, as highlighted by Cobirka et al. (2020). The prolonged duration of subclinical mastitis can facilitate the transmission of pathogens within the herd. Therefore, these findings emphasize the need for effective monitoring and management strategies to control subclinical mastitis and its impact on herd health and milk production. Further research and implementation of preventive measures are warranted to address this significant health concern in dairy herd.
The primary causative factors frequently encountered are bacteria, with over 150 Gram-positive and Gram-negative bacterial strains identified as mastitis pathogens. These bacteria can be categorized into contagious, transmitted from other infected quarters, and environmental, originating from the surrounding environment (Ashraf and Imran, 2020; Cobirka et al., 2020; Krishnamoorthy et al., 2021; Ndahetuye et al., 2019 and Ruegg, 2017).
The prevalence of antimicrobial resistance and genetic determinants in bacteria causing bovine mastitis is a growing concern in Algeria and worldwide. Several studies have investigated this issue, focusing on pathogens such as Staphylococcus aureus (S. aureus) and Escherichia coli (E. coli), Table 2.
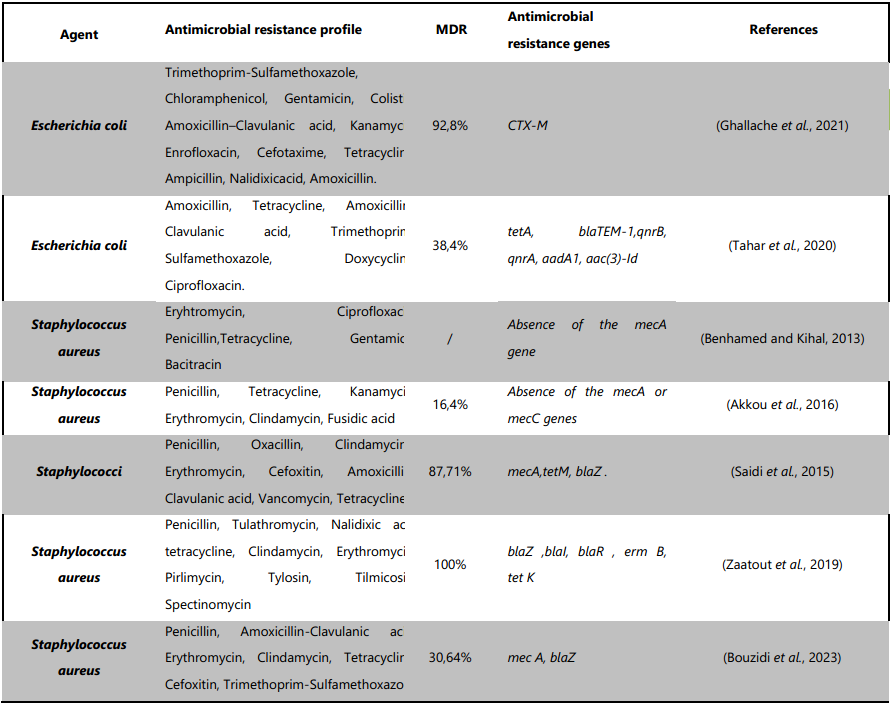
Table 2. Antibiotic susceptibility profiles and antimicrobial resistance genes detected in S. aureus and E. coli isolates from Algerian dairy cows. The data reveal widespread resistance, including methicillin-resistant S. aureus (MRSA) and multidrug-resistant (MDR) E. coli, with the presence of key resistance genes, highlighting concerns over antibiotic misuse and the need for improved surveillance and control measures.
S. aureus is a resilient bacterium that can survive in various environmental conditions, making it a major cause of intra-mammary infections in dairy cattle. The bacterium is highly prevalent in dairy farms, with its prevalence varying across different regions. The robust nature of S. aureus allows it to thrive in various settings, including dairy and beef production, processing, and supply lines. It is also known to be prevalent in raw meat, raw milk, equipment, and personnel who handle milk and meat. This widespread presence of S. aureus in the dairy farming environment emphasizes the need for stringent hygiene practices to prevent biofilm formation and the spread of infection within herds (Gizaw et al., 2023).
Akkou et al. (2016) reported S. aureus isolation in 74% of herds with 5–25 cows, offering valuable insights into mastitis epidemiology in smaller dairy farms. Furthermore, this pathogen was directly implicated in 29.8% of the cases, emphasizing its role as a primary contributor to the mastitis burden. The findings are consistent with other provincial studies in Algeria, such as the investigation conducted in Oran by Benhamed and Kihal (2013), which reported a notably high frequency of S. aureus in bovine mastitis. In this specific region, S. aureus was identified as the causative agent in 38.9% of both clinical and subclinical mastitis cases. These regional studies contribute to the broader understanding of the distribution and prevalence of S. aureus in different areas of the country. However, it is essential to approach the comparison of results from various prevalence studies with caution due to potential differences in sample selection and bacterial cultivation techniques (Akkou et al., 2016).
Several studies have emphasized the necessity of accurately identifying S. aureus infected cows to ensure the effective implementation of mastitis control programs (Akkou et al., 2016; Benhamed and Kihal, 2013; Bouzidi et al., 2023 and Zaatout et al., 2019).
Public health is seriously concerned when it comes to bacteria identified in milk that have genes for antibiotic resistance, especially when dairy farming is involved. The abuse or misuse of antibiotics in animal husbandry has the potential to promote the establishment of resistant diseases, especially in developing countries (Sipahi et al., 2023). Antibiotic-resistant bacteria not only threaten the health of diseased animals but also have the potential to impact healthy animals, which makes this issue especially pertinent in the case of bovine mastitis. Antibiotic resistance that makes it difficult to treat mastitis might cause dairy farmers to lose money and become less productive. Promoting appropriate antibiotic usage in the dairy business and increasing public knowledge of the effects of antimicrobial resistance are crucial in order to reduce this growing threat (Dong et al., 2022) and (Sipahi et al., 2023).
The veterinary clinics prolonged and frequent use of antibiotics like ampicillin, amoxicillin, cloxacillin, penicillin G, oxacillin, streptomycin and tetracycline has resulted in the emergence of resistant strains of bacteria (Saidi et al., 2015). Although little is known about the use of vancomycin in dairy cattle, the Saidi et al., 2015 study emphasizes the high frequency of 76.2% of vancomycin-resistant S. aureus isolates in dairy animals as well as the lack of vanA in study (Saidi et al., 2021). However, no research has been conducted elsewhere that reports the presence of vancomycin-resistant S. aureus in bovine mastitis in Algeria.
The most commonly antimicrobial resistance determined was against penicillin, erythromycin and tetracycline, Table2. The study of Saidi et al., 2015 conducted in Alger found that S. aureus isolates from bovine mastitis were highly resistant to penicillin (95.23%), oxacillin (80.95%), clindamycin (80.95%) and erythromycin (76.19%). MRSA strains were also detected, being methicillin-resistant and resistant to multiple antibiotics.
Almost the same results were reported by other authors (Akkou et al., 2016, Benhamed and Kihal, 2013b) especially on resistance to penicillin. The study conducted in Tiaret found that 30.64% of S. aureus isolates from subclinical bovine mastitis were multi-drug resistant, and 8% were MRSA isolates. The study also detected virulence genes, including eta, icaA, icaD, etb, luk E-D and sea, which play an important role in the pathogenesis of subclinical bovine mastitis (Bouzidi et al., 2023). Bouzidi's results brought to light the prevalence of several genetic components in MRSA and methicillin-susceptible S .aureus (MSSA) isolates that encode resistance to antibiotics. This may be the result of high antibiotic use in dairy cows, which promotes the selection of antibiotic-resistant strains, or it may be the result of mobile genetic elements encoding antibiotic resistance being exchanged (via conjugation, transposition and transduction) from resistant S. aureus strains (Bouzidi et al., 2023).
However, there were variations in the frequency of Multi-drug resistance phenotypes and resistance genes found in each study. In the study of Saidi et al., 2015, the distribution of antibiotic resistance genes was as mecA (100%) and tetM (100%) followed by blaZ (42.85%), whereas Bouzidi et al. in 2023, found that out of 59 penicillin-resistant isolates, 14 harbored the blaZ gene; one of them co-harbored the mecA gene. Genes conferring resistance to tetracycline (tet K), penicillin (blaZ, bla I and bla R) and macrolide–lincosamide–streptogramin B (erm B and ermA) were also detected in the S. aureus from the study of Zaatout et al. in 2019. However, no mecA genes were detected in the studies of Akkou et al. in 2016 and Benhamed and Kihal in 2013.
The blaZ encodes a protein penicillinase which is a type of beta-lactamase. This protein inactivates penicillin by hydrolyzing the beta-lactam ring, and it is responsible for beta-lactam resistance in Staphylococci. Staphylococcal beta-lactamase acts as an extended-spectrum beta-lactamase and contributes to borderline resistance to penicillin (Nomura et al., 2020). The high incidence of penicillin resistance in Staphylococci is largely due to the presence of the blaZ gene (Ferreira et al., 2016).
The mecA gene is responsible for encoding a penicillin-binding protein in methicillin resistant Staphylococcus spp. However, studies have shown that mecA may also promote resistance to other beta-lactam antibiotics (Abebe et al., 2020) and (Gueimonde et al., 2013). For this reason, The presence of the mecA gene along with the blaZ gene in bacteria is very important and poses a threat to public and animal health due to the constant evolution of drug resistance in bacteria (Andersson et al., 2020) and (Tóth et al., 2020). Different prevalences can be detected in studies conducted in different regions or in studies conducted in the same region at different times (Sipahi et al., 2023).
Furthermore, the occurrence of tetracycline-resistant and macrolide/clindamycin-resistant genes among bovine Staphylococci isolates has been observed in different countries, suggesting their role in promoting the evolution and development of antibiotic resistance (Mzee et al., 2023), (Saidi et al., 2019 and Zaatout et al., 2019).
Along with Staphylococci and Streptococci, E. coli is one of the most commonly isolated causal organisms linked to bovine intramammary infection (Burvenich et al., 2003). E. coli multidrug resistance is a concerning problem that is becoming more and more prevalent in both human and veterinary medicine globally.
Although, E. coli is naturally sensitive to practically all clinically important antibiotics, but, it is able to acquire resistance genes, mostly through horizontal gene transfer (Poirel et al., 2018). E. coli in milk is a cause for concern due to its association with fecal contamination (Kagkli et al., 2007).
In Algeria, the prevalence of E. coli in milk samples was 26.94% in Kolea and Birtouta Province (Ghallache et al., 2021) close to the prevalence 26% found in Medjana province, Sidi Embarak province, and El Achir region (Tahar et al., 2020). Indeed, mastitis frequently involves the bacteria E. coli (Goulart and Mellata, 2022). When it comes to the contamination of milk with E. coli, farm hygiene is a major factor.
The prevalence of environmental E. coli may be associated with poor farm cleanliness and poor slope of stable areas. Feces which are common sources of E. coli can contaminate the premises directly or indirectly through bedding, calving stalls, udder wash water, and milker’s hands (Abutarbush, 2010)
It is imperative to follow proper hygiene procedures, such as cleaning the udders before milking to minimizing bacterial contaminations (Seferoğlu and Kirkan, 2022). Both human digestive tracts and those of numerous animal species used in food production contain E. coli. The genetic flexibility and adaptability of this bacteria to constantly changing environments allows it to acquire a great number of antimicrobial resistance mechanisms. New strains of multidrug-resistant foodborne bacteria, such E. coli that produces extended spectrum beta-lactamase (ESBL), have been linked to the introduction of new types of resistant bacteria in food-producing animals due to the usage of antibiotics that are crucial for human medicine (Ramos et al., 2020).
E. coli in cows with clinical mastitis in 42 different dairy farms have been investigated. The most frequently observed resistance was to amoxicillin 86.5%, tetracycline 75%, amoxicillin–clavulanic acid 59.6%, trimethoprim-sulfamethoxazole 36.5%, doxycycline 13.5%, and ciprofloxacin 13.5%. Multidrug resistance was observed in 38.4% of isolates (Tahar et al., 2020). In a similar study, in the same country for a similar survey, the antibiotic resistance profile of the isolated E. coli strains from subclinical mastitis was to amoxicillin 75.3%, nalidixic acid 74.3%, ampicillin 57.7%, tetracycline 52%, cefotaxime 51.5%,enrofloxacin 39.2%, kanamycin 31.9%, amoxicillin/clavulanate 23.3%, colistin 13.4%, gentamicin 12.3%, chloramphenicol 6.2% and 3.1% of the strains were resistant to trimethoprim-sulfamethoxazole. Furthermore, most of the E. coli strains (92.8%) were resistant to more than one antibiotic (Ghallache et al., 2021). The extensive usage of amoxicillin, tetracycline, and amoxicillin-clavulanic acid in veterinary medicine, along with their affordability, are the primary causes of the elevated resistance levels seen in the current investigations (Tahar et al., 2020).
Whereas, following the results of these studies the best antibiotics in the fight against mastitis in Algeria are gentamicin and the combination trimethoprim-sulfamethoxazole. The effectiveness of enrofloxacin therapies has been shown in other experimental or clinical investigations (Persson et al., 2015).
Resistance to beta-lactams is possibly the most found in E. coli isolates from bovine mastitis (Naranjo-Lucena and Slowey, 2023). Therefore, the observed resistance rate in the studies was quite high indicating that E. coli strains producing beta-lactamases were common in Algeria. The blaTEM-1 gene (30.7%) was only present in the ESBL-producing E. coli isolates that were detected in the study of Tahar and colleagues in 2020. The bla CTX-M has also been reported in another study from 69.2% of the E. coli strains where 78% of the bacteria that were resistant to CTX displayed an ESBL-classic synergy pattern (Ghallache et al., 2021).
The blaTEM-1 and CTX-M genes are significantly present in these isolates, indicating a troubling level of resistance to third-generation cephalosporins. These data further emphasize the frequency of ESBL-producing E. coli.
The presence of blaTEM and blaCTX-M types in Enterobacteriaceae isolates from cows is worrisome, as ESBL E. coli producers, particularly the CTX-M type, play a significant role in causing human infections, both in healthcare settings and within the community. This raises concerns about the potential transmission and impact on public health (De Angelis et al., 2020).
E. coli resistant to beta-lactams poses a significant concern because these isolates were not only resistant to beta-lactams but also revealed genes encoding resistance to tetracycline, quinolones and aminoglycosides. Furthermore, the simultaneous occurrence of resistance genes for tetracyclines, quinolones and aminoglycosides alongside beta-lactams resistance genes in multidrug-resistant (MDR) E. coli suggests a common localization of these genes on mobile genetic elements (Tahar et al., 2020).
Tetracycline is a wide-spectrum bacteriostatic antibiotic used in cattle to treat various infections. However, its misuse has prompt some countries, including European countries, to prohibit its use in animal feed (European Commission, 2019). National studies have shown high rates of tetracycline resistance in E. coli with 52% (Ghallache et al., 2021) and 75% (Tahar et al., 2020), which suggests that this issue is widespread with previous reports from Algeria. tetA was the most prevalent tetracycline gene, identified, followed by tetB and tetC genes. Among the isolates resistant to ciprofloxacin, two quinolone genes were detected: qnrB and qnrA.
The examination for genes encoding aminoglycoside-modifying enzymes revealed that the isolates carried the aadA1 and aac(3)-Id genes. Fortunately, none of the tested isolates contained the tetJ, aph(30), aac(60), ant, armA, and aac(60)-Ib cr genes (Tahar et al., 2020).
Resistance to colistin that is of particular importance, have also been detected in 13.4% of the E. coli strains (Ghallache et al., 2021). Colistin is currently recognized as the final option for treating infections caused by multidrug-resistant (MDR) Gram-negative bacteria (Mohapatra et al., 2021). It is a widely used antibiotic for enterobacterial infections and growth promotion in veterinary medicine and agriculture. However, the rapid proliferation of mcr resistance genes has been aided by this broad usage. Finding the prevalence of mcr resistance genes in these reservoirs is crucial for improving the implementation of control and prevention efforts as well as for gaining a more accurate knowledge of the global expansion. Therefore, it would be interesting to look at the plasmid profiles of isolated strains in order to determine the genetic basis of colistin resistance (Kumar et al., 2020).
Conclusion
The study reveals a high prevalence of S. aureus and E. coli in Algerian dairy cows, both of which exhibit significant antibiotic resistance. Concerns about poor hygiene standards and the misuse of antibiotics on dairy farms are raised by the occurrence of antibiotic-resistant strains and subclinical mastitis in cows. Notably, bacteriological and antimicrobial resistance studies of bovine mastitis have not been fully performed in Algeria, particularly in terms of limited molecular detection of resistance genes. Furthermore, the research suggests that veterinarians and herd managers need to pay close attention to the increasing number of methicillin-resistant S. aureus and multidrug-resistant E. coli isolates found in Algerian dairy farms. The findings also highlight the importance of developing strategic plans for the treatment, prevention, and control of mastitis in Algeria, as well as the need for further research to evaluate virulence factors and to conduct whole genome sequencing to characterize the virulence and resistance genes of the isolates. The study's results can be used as a baseline for future investigations and to inform treatment policies and antimicrobial strategies.
References
🕮 Abebe, E; Gugsa, G and Ahmed, M (2020). Review on Major Food-Borne Zoonotic Bacterial Pathogens. J Trop Med 2020, PP : 1-19.
🕮 Abutarbush, S.M (2010). Veterinary Medicine - A Textbook of the Diseases of Cattle, Horses, Sheep, Pigs and Goats, 10th edition. Can Vet J 51 : 541.
🕮 Akkou, M ; Antri, K ; Bachtarzi, M.-A ; Bes, M ; Tristan, A ; Dauwalder, O ; Kaidi, R ; Meugnier, H ; Tazir, M and Etienne, J (2016). Phenotypic and genotypic characterization of Staphylococcus aureus strains associated with bovine mastitis and nasal carriage of workers in contact to animals in Algeria. Pak. Vet. J., 36 : 184-188.
🕮 Andersson, D.I ; Balaban, N.Q ;Baquero, F ; Courvalin, P ;Glaser, P ; Gophna, U ; Kishony, R ; Molin, S and Tønjum, T (2020). Antibiotic resistance: turning evolutionary principles into clinical reality. FEMS Microbiology Reviews., 44 :171-188.
🕮 Ashraf, A ; Imran, M (2020). Causes, types, etiological agents, prevalence, diagnosis, treatment, prevention, effects on human health and future aspects of bovine mastitis. Anim. Health. Res. Rev., 21:36-49.
🕮 Benhamed, N and Kihal, M (2013). Biodiversity of molecular profile of Staphylococcus aureus isolated from bovine mastitis cases in West Algeria. J. Bacteriol. Res., PP : 541-45.
🕮 Bouzidi, S ; Bourabah, A ; Cheriet, S ; Abbassi, M.S ; Meliani, S and Bouzidi, H (2023). Occurrence of virulence genes and methicillin-resistance in Staphylococcus aureus isolates causing subclinical bovine mastitis in Tiaret area, Algeria. Letters in Applied Microbiology., 76 :1-9.
🕮 Burvenich, C ; Van Merris, V. ; Mehrzad, J ; Diez-Fraile, A ; Duchateau, L (2003). Severity of E. coli mastitis is mainly determined by cow factors. Vet. Res., 34 : 521-564.
🕮 Cobirka, M ; Tancin, V and Slama, P (2020). Epidemiology and Classification of Mastitis. Animals., 10 :2212.
🕮 Regulation, EU (2019). Commission Regulation (EU) 2019/4 of the European Parliament and of the Council of 11 December 2018 on the manufacture, placing on the market and use of medicated feed, amending Regulation (EC) No 183/2005 of the European Parliament and of the Council and repealing Council Directive 90/167/EEC (Text with EEA relevance). Off. J. Eur. Union., 4 :1-23.
🕮 De Angelis, G ; Del Giacomo, P ; Posteraro, B ; Sanguinetti, M and Tumbarello, M (2020). Molecular Mechanisms, Epidemiology, and Clinical Importance of β-Lactam Resistance in Enterobacteriaceae. International Journal of Molecular Sciences., 21 : 5090.
🕮 Dong, L ; Meng, L ; Liu, H ; Wu, H ; Schroyen, M ; Zheng, N and Wang, J (2022). Effect of Cephalosporin Treatment on the Microbiota and Antibiotic Resistance Genes in Feces of Dairy Cows with Clinical Mastitis. Antibiotics (Basel)., 11 :117.
🕮 Ferreira, A.M ; Martins, K.B ; Silva, V.R. da ; Mondelli, A.L and Cunha, M. de L.R. de S. da (2016). Correlation of phenotypic tests with the presence of the blaZ gene for detection of beta-lactamase. Braz J Microbiol .,48 :159-166.
🕮 Ghallache, L ; Mohamed-Cherif, A ; China, B ; Mebkhout, F ; Boilattabi, N ; Bouchemal, A ; Rebia, A ; Ayachi, A ; Khelef, D and Miroud, K (2021). Antibiotic Resistance Profile of Escherichia coli Isolated from Bovine Subclinical Mastitis of Dairy Farms in Algeria from 2017 to 2019. World’s Veterinary Journal., 11 :402-415.
🕮 Gizaw, F ; Kekeba, T ; Teshome, F ; Kebede, M ; Abreham, T ; Berhe, H.H ; Ayana, D ; Edao, B.M ; Waktole, H ; Tufa, T.B ; Abunna, F ; Beyi, A.F ; Abdi, R.D (2023).Multidrug-Resistant Staphylococcus aureus Strains Thrive in Dairy and Beef Production, Processing, and Supply Lines in Five Geographical Areas in Ethiopia. Vet Sci., 10 :663.
🕮 Goulart, D.B and Mellata, M (2022). Escherichia coli Mastitis in Dairy Cattle: Etiology, Diagnosis, and Treatment Challenges. Front. Microbiol., 13 : 928346.
🕮 Gueimonde, M ; Sánchez, B ; G. De Los Reyes-Gavilán, C and Margolles, A (2013). Antibiotic resistance in probiotic bacteria. Front. Microbiol., 4 :202.
🕮 Kagkli, D.M ; Vancanneyt, M ; Vandamme, P ; Hill, C and Cogan, T.M (2007). Contamination of milk by enterococci and coliforms from bovine faeces. Journal of Applied Microbiology., 103 :1393-1405.
🕮 Krishnamoorthy, P ; Suresh, K.P ; Jayamma, K.S ; Shome, B.R ; Patil, S.S and Amachawadi, R.G (2021). An Understanding of the Global Status of Major Bacterial Pathogens of Milk Concerning Bovine Mastitis: A Systematic Review and Meta-Analysis (Scientometrics). Pathogens., 10 : 545.
🕮 Kumar, H ; Chen, B.-H ; Kuca, K ; Nepovimova, E ; Kaushal, A ; Nagraik, R ; Bhatia, S.K ; Dhanjal, D.S ; Kumar, V ; Kumar, A ; Upadhyay, N.K ; Verma, R and Kumar, D (2020). Understanding of Colistin Usage in Food Animals and Available Detection Techniques: A Review. Animals (Basel)., 10 :1892.
🕮 Mohapatra, S.S ; Dwibedy, S.K and Padhy, I (2021). Polymyxins, the last-resort antibiotics: Mode of action, resistance emergence, and potential solutions. J Biosci.,46 :85.
🕮 Mzee, T ; Kumburu, H ; Kazimoto, T ; Leekitcharoenphon, P ; van Zwetselaar, M ; Masalu, R ; Mlaganile, T ; Sonda, T ; Wadugu, B ; Mushi, I ; Aarestrup, F.M and Matee, M (2023). Molecular Characterization of Staphylococcus aureus Isolated from Raw Milk and Humans in Eastern Tanzania: Genetic Diversity and Inter-Host Transmission. Microorganisms., 11 :1505.
🕮 Naranjo-Lucena, A and Slowey, R (2023). Invited review: Antimicrobial resistance in bovine mastitis pathogens: A review of genetic determinants and prevalence of resistance in European countries. Journal of Dairy Science., 106 : 1-23.
🕮 Ndahetuye, J.B ; Persson, Y ; Nyman, A.-K ; Tukei, M ; Ongol, M.P and Båge, R (2019). Aetiology and prevalence of subclinical mastitis in dairy herds in peri-urban areas of Kigali in Rwanda. Trop Anim Health Prod., 51:2037-2044.
🕮 Nomura, R ; Nakaminami, H ; Takasao, K ; Muramatsu, S ; Kato, Y ; Wajima, T and Noguchi, N (2020). A class A β-lactamase produced by borderline oxacillin-resistant Staphylococcus aureus hydrolyses oxacillin. Journal of Global Antimicrobial Resistance., 22 :244-247.
🕮 Persson, Y ; Katholm, J ; Landin, H and Mörk, M.J (2015). Efficacy of enrofloxacin for the treatment of acute clinical mastitis caused by Escherichia coli in dairy cows. Veterinary Record., 176 : 673-673.
🕮 Ramos, S ; Silva, V ; Dapkevicius, M. de L.E ; Caniça, M ; Tejedor-Junco, M.T ; Igrejas, G and Poeta, P (2020). Escherichia coli as Commensal and Pathogenic Bacteria among Food-Producing Animals: Health Implications of Extended Spectrum β-Lactamase (ESBL) Production. Animals (Basel).,10 :2239.
🕮 Rana, E.A ; Fazal, M.A and Alim, M.A (2022). Frequently used therapeutic antimicrobials and their resistance patterns on Staphylococcus aureus and Escherichia coli in mastitis affected lactating cows. Int J Vet Sci Med., 10 :1-10.
🕮 Ruegg, P.L (2021). What Is Success? A Narrative Review of Research Evaluating Outcomes of Antibiotics Used for Treatment of Clinical Mastitis. Front. Vet. Sci.,8 :639641.
🕮 Ruegg, P.L (2017). A 100-Year Review: Mastitis detection, management, and prevention. Journal of Dairy Science., 100 :10381-10397.
🕮 Saidi, R ; Cantekin, Z ; Khelef, D ; Ergün, Y ; Solmaz, H and Kaidi, R (2015). Antibiotic Susceptibility and Molecular Identification of Antibiotic Resistance Genes of Staphylococci Isolated from Bovine Mastitis in Algeri. KafKas Universitesi veteriner faKUltesi Dergisi., 21.
🕮 Saidi, R ; Cantekin, Z ; Mimoune, N ; Ergun, Y ; Solmaz, H ; Khelef, D and Kaidi, R (2021). Investigation of the presence of slime production, VanA gene and antiseptic resistance genes in Staphylococci isolated from bovine mastitis in Algeria. Veterinarska stanica., 52 : 0-0.
🕮 Saidi, R ; Mimoune, N ; Baazizi, R ; Benaissa, M.H ; Khelef, D and Kaidi, R (2019). Antibiotic susceptibility of Staphylococci isolated from bovine mastitis in Algeria. J Adv Vet Anim Res., 6 :231-235.
🕮 Seferoğlu, Y and Kirkan, Ş (2022). Bovine Escherichia coli Mastitis and Effects on Milk Microbiota. Animal Health Production and Hygiene., 11 :56-65.
🕮 Sipahi, N; Kaya, E ; Çelik, C and Pınar, O (2023). The Characterization and Beta-Lactam Resistance of Staphylococcal Community Recovered from Raw Bovine Milk. Antibiotics.,12 :556.
🕮 Tahar, S ; Nabil, M.M ; Safia, T ; Ngaiganam, E.P ; Omar, A ; Hafidha, C ; Hanane, Z ; Rolain, J.-M and Diene, S.M (2020). Molecular Characterization of Multidrug-Resistant Escherichia coli Isolated from Milk of Dairy Cows with Clinical Mastitis in Algeria. Journal of Food Protection., 83 :2173-2178.
🕮 Tommasoni, C ; Fiore, E ; Lisuzzo, A and Gianesella, M (2023). Mastitis in Dairy Cattle: On-Farm Diagnostics and Future Perspectives. Animals (Basel)., 13 : 2538.
🕮 Tóth, A.G ; Csabai, I ; Krikó, E ; Tőzsér, D ; Maróti, G ; Patai, Á.V ; Makrai, L ; Szita, G and Solymosi, N (2020). Antimicrobial resistance genes in raw milk for human consumption. Sci Rep., 10 : 7464.
🕮 Zaatout, N ; Ayachi, A ; Kecha, M and Kadlec, K (2019). Identification of staphylococci causing mastitis in dairy cattle from Algeria and characterization of Staphylococcus aureus. J Appl Microbiol., 127 : 1305-1314.